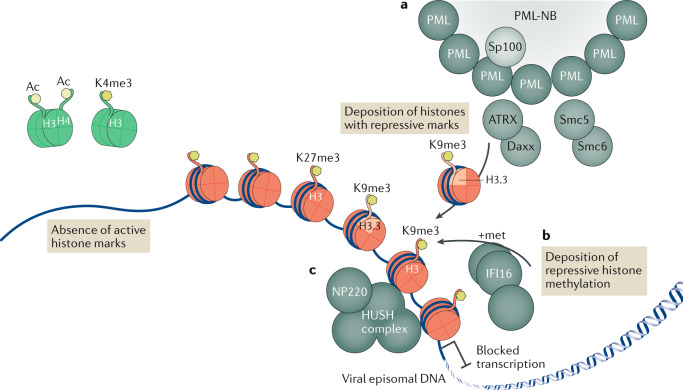
Fig. 1. Epigenetic repression of viral DNA.
Upon entry into the cell nucleus, the DNA of many viruses initiates the replication process adjacent to subnuclear structures called pro-myelocytic leukaemia nuclear bodies (PML-NBs). However, PML-NBs are an aggregation site for many heterochromatic repression proteins, which load repressive heterochromatin onto viral DNA that shuts down viral transcription. In the absence of viral de-repression factors, viral episomal DNA lacks active histone marks (shown as green histones with the active marks H3Ac, H4Ac and H3K4me3). a | Several PML-NB components — including PML itself, Sp100, Smc5, Smc6, Daxx and ATRX — are involved in the epigenetic repression of viruses, with the Daxx–ATRX complex having specifically been found to load the histone variant H3.3 bearing repressive marks onto viral DNA, leading to the accumulation of heterochromatic marks and blocked transcription (DNA is shown associated with red histones with the repressive marks H3K9me3 and H3K27me3). b | The innate immune DNA sensor IFI16 drives an alternative mechanism of antiviral epigenetic repression, promoting repressive methylations on histone tail H3K9. c | Specifically for the retrovirus murine leukaemia virus, NP220 and the human silencing hub (HUSH) complex have also been shown to deposit heterochromatic marks on unintegrated viral DNA.