Abstract
Background
Short non-coding microRNAs (miRNAs) are involved in various cellular processes during disease progression of Crohn’s disease (CD) and remarkably stable in feces, which make them attractive biomarker candidates for reflecting intestinal inflammatory processes. Here we investigated the potential of fecal miRNAs as noninvasive and translational CD biomarkers.
Methods
MiRNAs were screened in feces of 52 patients with CD and 15 healthy controls using RNA sequencing and the results were confirmed by PCR. The relationship between fecal miRNA levels and the clinical CD activity index (CDAI) or CD endoscopic index of severity (CDEIS) was explored, respectively. Additionally, fecal miRNAs were investigated in dextran sodium sulfate, adoptive T-cell transfer, and Helicobacter typhlonius/stress-induced murine colitis models using the NanoString platform.
Results
Nine miRNAs (miR-15a-5p, miR-16-5p, miR-128-3p, miR-142-5p, miR-24-3p, miR-27a-3p, miR-223-3p, miR-223-5p, and miR-3074-5p) were significantly (adj. P < 0.05, >3-fold) increased whereas 8 miRNAs (miR-10a-5p, miR-10b-5p, miR-141-3p, miR-192-5p, miR-200a-3p, miR-375, miR-378a-3p, and let-7g-5p) were significantly decreased in CD. MiR-192-5p, miR-375, and miR-141-3p correlated (P < 0.05) with both CDAI and CDEIS whereas miR-15a-5p correlated only with CDEIS. Deregulated expression of miR-223-3p, miR-16-5p, miR-15a-5p, miR-24-3p, and miR-200a-3p was also observed in murine models. The identified altered fecal miRNA levels reflect pathophysiological mechanisms in CD, such as Th1 and Th17 inflammation, autophagy, and fibrotic processes.
Conclusions
Our translational study assessed global fecal miRNA changes of patients with CD and relevant preclinical models. These fecal miRNAs show promise as translational and clinically useful noninvasive biomarkers for mechanistic investigation of intestinal pathophysiology, including monitoring of disease progression.
Keywords: Crohn’s disease, miRNAs, biomarker, noninvasive, colitis model
INTRODUCTION
Crohn’s disease (CD) is a chronic, relapsing, and remitting disorder within the gastrointestinal tract.1 The pathogenesis is characterized by a multilayered interplay of genetic susceptibility, environmental triggers, intestinal dysbiosis, and disturbed immune homeostasis resulting in intestinal inflammation.2,3 However, the exact molecular pathophysiology underlying the mechanism of CD is not completely understood to date. The clinical status is measured using clinical indexes, such as the most commonly used Crohn’s disease activity index (CDAI) and the extent of disease can be graded following ileocolonoscopy according to the Crohn’s disease endoscopic index of severity (CDEIS). This invasive procedure is complicated and time-consuming making these modalities unsuitable for frequent monitoring of patients with CD.4 Serum C-reactive protein and fecal calprotectin are the most commonly used biomarkers although they represent general and unspecific pro-inflammatory markers. Therefore, there is an unmet need to identify biomarkers, which reflect molecular mechanisms during early disease development and progression and potentially allow monitoring of predictive and/or prognostic treatment effects.
MicroRNAs (miRNAs) are small, non-coding RNA molecules that are 18–23 nucleotides in length and critical regulators of cell differentiation, growth, apoptosis, and proliferation. They hereby function as regulators of protein synthesis by either inducing mRNA degradation or repressing translation.5 Recent studies uncovered distinct miRNA profiles in the intestinal tissue from patients with CD.6–8 Functional studies revealed that deregulated miRNA expression resulted in altered Th17 immune response, intestinal epithelial barrier integrity, and autophagy, which are hallmarks of CD pathogenesis. Interleukin-23 (IL-23) mediated Th17 inflammation is modulated by miR-10a via targeting IL-12/IL-23p40 expression. The expression of miR-10a was markedly decreased in inflamed mucosal tissue of patients with CD compared to healthy controls (HC), while IL-12/IL-23p40 was significantly increased.9 In particular, IL-23 regulated miR-223-3p expression was increased in mucosal tissue from patients with CD and animal models of intestinal inflammation,10–12 which in turn negatively targeted claudin-8, a critical member in the maintenance of normal intestinal barrier integrity.13 The intestinal expression of nucleotide-binding oligomerization domain-containing protein 2 (NOD2), an important regulator of intestinal barrier integrity, inflammatory, and autophagy processes, was increased in patients with CD,14 whereas the expression of its regulating miR-192 was decreased in inflamed colon mucosa of patients with CD.15
Circulating cell-free miRNAs can be detected in a wide variety of body fluids including blood and feces. Recent studies revealed differentially expressed miRNAs in peripheral blood of patients with CD compared to HC.16,17 Unfortunately, miRNA expression analyses in blood predominantly reflect the extrusion of miRNAs from distant tissues or organs. Moreover, there was just a little overlap of miRNA expression profiles in blood and mucosal tissue and a poor correlation with disease activity.17,18 Compared to blood biomarkers, fecal biomarkers have the advantage of increased specificity for inflammatory processes localized to the bowel as colonocytes are continuously released into the intestinal lumen.19
In contrast to the most commonly used inflammatory fecal biomarkers calprotectin and lactoferrin, miRNAs have the potential to reflect molecular pathophysiological mechanisms during disease progression. Furthermore, miRNAs are remarkably stable in stool and could be reproducibly detected even after long-term storage of fecal samples for many years.20,21 It was previously shown that a panel of fecal miRNAs could differentiate patients with colorectal cancer (CRC) from HC and identify different TNM stages with high sensitivity and specificity.22 Recently, there have been first attempts to evaluate the potential of fecal miRNAs as inflammatory bowel disease biomarkers. However, these studies were limited to targeted approaches using a few selected miRNAs or small screening cohorts (6 donors per group) and predefined miRNA panels.23–25
In the present study, we performed unbiased RNA sequencing to investigate global fecal miRNA profiles in patients with CD and demonstrate that robustly expressed fecal miRNAs most likely represented the mucosal miRNA expression. We also show that fecal miRNA profiles differentiated patients with CD from HC and that distinct fecal miRNAs significantly correlated with CDAI and CDEIS scores. Moreover, changes in fecal miRNA levels provided insights into pathophysiological mechanisms involved in human CD. We further expanded the concept of fecal miRNA profiling to mouse models of dextran sodium sulfate (DSS), adoptive T-cell transfer, and Helicobacter typhlonius/stress-induced intestinal inflammation. In this, to our knowledge, most comprehensive translational study, we provide evidence that diseased animals also exhibit an altered fecal miRNA profile including several miRNAs that translate back to human CD. Thus, fecal miRNAs show promise as translational and clinically useful, noninvasive biomarkers for investigation of intestinal pathophysiology in CD.
MATERIALS AND METHODS
Subjects and Ethical Considerations
In total 52 adult patients suffering from mild to severe active CD with a CDEIS score greater than 4 were recruited in a sub-study of clinical phase II study NCT02031276 (Table 1). All included patients had a diagnosis of CD (16× ileocolonic, 10× colonic, 10× ileal, 16× disease location not available) at least 3 months before the start of the study. The presence of mucosal ulcers in the ileum, colon, or both intestinal segments was confirmed by ileocolonoscopy. CD Patients with stenoses, strictures, short gut syndrome, current colostomy, or ileostomy were excluded from this study. Patients with CD who underwent intra-abdominal surgeries not more than 3 months and bowel diversion or resection not more than 6 months before study start were also excluded. Patients were included if they were treatment-naive or experienced to one or more TNF antagonists (infliximab, adalimumab, or certolizumab pegol) at a dose approved for CD. However, anti-TNF therapy needed to be stopped minimum 8 weeks prior to the study start. Patients who received T- or B-cell depleting agents, natalizumab or efalizumab, within 6 months before study start or were treated with ustekinumab were excluded from the study.
Table 1.
Clinical Characteristics of Patients with Crohn’s Disease and Healthy Controls
| Crohn’s Disease | Healthy Controls | |
|---|---|---|
| Number of subjects | 52 | 15 |
| Male percentage | 25.0 | 53.3 |
| Mean age (years, range) | 38 (19–74) | 38 (23–65) |
| Mean CDEIS (range)a | 13.3 ± 6.2 (5.2–35.0) | — |
| Mean CDAI (range)b | 299.0 ± 76.7 (143.8–418.7) | — |
CDEIS, Crohn’s disease endoscopic index of severity; CDAI, Crohn’s disease activity index.
aObtained from 29 patients with Crohn’s disease.
bObtained from 33 patients with Crohn’s disease.
Out of the 52 patients with CD, 11 patients provided only ileum biopsies, 11 patients only colon biopsies, 22 patients both ileum and colon biopsies, and 8 patients no biopsies. Moreover, 1 subject out of the 22 donated an additional ileal and colonic biopsy, respectively. In total, 68 intestinal CD biopsies were available. The control group consisted of 15 age-matched HC (Table 1). The control subjects had no diagnosis of functional gastrointestinal disorders, no abnormal stool or bowel frequency, and no abnormal stool consistency. The institutional review board or ethics committee of each participating center approved the study protocol and all donors were informed about the study and gave informed consent.
Naturally evacuated stool was collected by all 52 patients with CD and all 15 HC and stored at −80°C after delivery. Tissue biopsies were collected from inflamed regions of the respective intestinal segment from most patients with CD but no HC. Biopsies were placed in RNAlater (Ambion, Thermo Fisher Scientific, Waltham, MA), kept at 4°C for at least 24 hours, and then stored at −80°C.
Animal Models
Mice and ethical considerations
Animal studies conform to the guidelines of the “Guide for the Care and Use of Laboratory Animals” by the National Academy of Sciences. All experiments of the DSS (No. 13–019) and social stress (No. 1136) models were approved by the local authorities (Regierungspräsidium Tübingen, Germany) and the T-cell transfer model was approved by Boehringer Ingelheim’s Institutional Animal Care and Use Committee.
Mice of the DSS and adoptive T-cell transfer models were allowed to acclimatize for 2 weeks whereas mice of the social stress model were acclimatized for 1 week after delivery. Mice were housed with 12 hours light/dark cycle with access to food and water provided ad libitum.
DSS model
On day 0, 2% (wt/vol) DSS (molecular weight 36,000–50,000, MP Biomedicals, Santa Ana, CA) was added for 5 days to the drinking water of 12 weeks old specific pathogen-free (SPF) female C57BL/6NCrl mice (Charles River Laboratories, Sulzfeld, Germany). Control mice received no DSS. On day 5, DSS was removed and replaced with regular tap water. Mice were killed by cervical dislocation on day 6, 13, and 21 post-initiation of DSS-induced colitis. The control cohort comprised 4 animals on day 6 and 13 and 5 animals on day 21. DSS-exposed groups were represented by 3 animals on day 6 and 5 animals on days 13 and 21, respectively. Fresh fecal pellets were collected per animal, snap-frozen in liquid nitrogen, and then stored at −80°C until processed for RNA isolation.
Adoptive T-cell transfer model
Naive T-cell mediated colitis was induced in female CB.17 SCID recipient mice while CB6F1 female mice served as cell donors (Jackson Labs, Sacramento, CA). CB6F1 mice were humanely killed and spleens collected on ice. Following homogenization of spleens and RBC lysis, CD4+ cells were enriched from pooled spleen cells using a commercially available negative selection kit (Stemcell Technologies, Vancouver, BC) following the manufacturer’s protocol. These CD4+ enriched cells were stained with Alexa Flour488 conjugated anti-CD4 clone RM4-5, Alexa Flour647 conjugated anti-CD45RB clone C363-16A, and PE-conjugated anti-CD25 clone PC61 (eBioscience, San Diego, CA). CD4+ CD45RBhigh cells were sorted on a FACS Aria II (BD Biosciences, San Jose, CA). Nine recipient CB.17 SCID mice were injected intraperitoneally with 5 × 105 purified CD4+ CD45RBhigh donor lymphocytes in 200 μL of PBS whereas 9 naive animals served as a control. Fresh fecal pellets were collected at week 4 post-adoptive transfer and snap-frozen in liquid nitrogen. Samples were stored at −80°C until processed for RNA isolation.
Social stress model
Male SPF C57BL/6N (experimental mice, weighing 19–22 g) and male SPF CD-1 (dominant aggressor mice, weighing 30–35 g) mice were obtained from Charles River (Sulzfeld, Germany). After delivery mice were individually housed under SPF conditions until the start of the chronic psychosocial stress procedure. Prior to the start of the experiment, dominant resident CD-1 mice were exposed to bedding and stool pellets from H. typhlonius-positive mice until their own fecal samples were tested positive for H. typhlonius by PCR.
Chronic psychosocial stress was induced by the chronic subordinate colony (CSC) housing paradigm. Briefly, a group of 4 CSC mice was housed together with a dominant male CD-1 mouse for 19 consecutive days. To avoid habituation, CSC mice were placed in the home cage of a novel dominant aggressor male CD-1 mouse on days 8 and 15. Single-housed control mice were used as controls and remained undisturbed for the duration of the experiment, except for changing the bedding once a week and weighing. In the morning of day 20 single-housed control (n = 12) and CSC (n = 12) mice were euthanized by rapid decapitation following brief CO2 exposure. Colonic fecal samples were collected and frozen at −80°C until processed for RNA isolation.
MiRNA Isolation
Frozen feces were transferred into 1 mL of QIAzol Lysis Reagent (Qiagen, Hilden, Germany) and homogenized by a Precellys 24 homogenizer (Bertin Technologies, Rockville, MD) for 30 seconds at 6500 rpm using the Precellys Ceramic 1.4/2.8 mm beads Kit (Bertin Technologies). The homogenate was mixed with 300 µL of 1-bromo-3-chloropropane (Sigma-Aldrich, St. Louis, MO) and miRNAs were isolated using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Additionally, depletion of small fragments was performed using an Oligo Clean & Concentrator spin column (Zymo Research, Irvine, CA) according to the manufacturer’s protocol for human samples. The purified and depleted RNA samples were quantified by a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
MiRNAs from ileal and colonic tissue biopsies of the patients with CD were purified using the RNeasy Fibrous Tissue Mini Kit (Qiagen) as described by the manufacturer. Tissues were homogenized in 600 µL of RLT buffer for 20 seconds at 6500 rpm by a Precellys 24 homogenizer (Bertin Technologies) using the Precellys Steel 2.8 mm Kit (Bertin Technologies).
Small RNA Sequencing
MiRNA profiles in human feces and intestinal biopsies were investigated by small RNA sequencing (small RNA-seq) whereas fecal miRNA levels of murine model systems were determined by NanoString nCounter measurement.
Library preparation and sequencing
CleanTag Ligation Kit for Small RNA Library Prep (Trilink Biotechnologies, San Diego, CA) was used to prepare small RNA-seq libraries from 100 ng of intestinal total RNA and 10 ng of fecal total RNA according to the manufacturer’s protocol. Fifteen and 18 PCR cycles were run to amplify the sequencing constructs during library preparation of the intestinal and fecal samples, respectively. For the sequencing reaction, small RNA-seq library samples were spiked into library samples prepared for conventional RNA sequencing and clusters were generated by means of a cBot (Illumina, San Diego, CA) and TruSeq SR Cluster Kit v3-cBot-HS (Illumina). Samples were then sequenced on a HiSeq 2000 platform (Illumina) using the TruSeq SBS Kit v3-HS (Illumina). During single read sequencing, 52 nucleotides were sequenced per fragment. On average 10.9 million reads were sequenced per intestinal biopsy sample and 19.5 million reads per fecal sample.
Processing of small RNA-seq data
First, Minion, which is part of the Kraken pipeline, was used to infer 3′ adaptor sequences with Minion search adaptor. Adaptors identified by Minion were cross-referenced via the sibling program swan using a FASTA file of known Illumina adaptor sequences. Identified adaptor sequences were trimmed via Reaper and trimmed reads were mapped against the human reference genome hg19 using STAR version 2.3.0. Reads 16–26 nucleotides in length were filtered to quantify mature miRNAs and reads mapping to the miRNAs annotated in miRBase release 20 were counted applying featureCounts. Read counts of each mature miRNA per sample were normalized to counts per million miRNA read counts (cpm) and voom transformed using the functions calcNormFactors and voom, which are part of the linear models for microarray data (limma) software package version 3.26.3.
Differential expression analysis
Differential expression was calculated applying the linear models approach incorporated in the limma software package using Tibco Spotfire version 6.5.2 (TIBCO Software, Palo Alto). MiRNAs included in the biopsy miRNA set and fecal miRNA core set were considered for differential expression analysis of intestinal tissue biopsies and fecal samples, respectively.
MiRNAs greater than 3-fold differentially expressed between the compared groups with a P-value adjusted for multiple testing less than 0.05 (Benjamini–Hochberg procedure) were considered significantly differentially expressed. Tibco Spotfire (TIBCO Software) software was used to generate heatmaps and calculate principal component analyses. Hierarchical clustering was calculated using the unweighted pair group method with arithmetic mean as the default clustering method and City block distance measure.
NanoString nCounter Analysis
Sample preparation and data processing
Fecal miRNA profiles of the murine model systems were analyzed by the Mouse v1.5 nCounter miRNA Assay Kit (NanoString Technologies, Seattle, WA). Five hundred nanograms of fecal total RNA were used to process the samples as per manufacturers’ instructions. In brief, miRNAs ligated to miRNA-specific tags were incubated with probe sets overnight at 65°C for approximately 20 hours. Samples were then processed by the nCounter Prep Station and 555 fields of view were scanned on the nCounter Digital Analyzer (NanoString Technologies) to determine the miRNA counts per sample.
Count data were processed by the nSolver v3.0 software (NanoString Technologies) to subtract background noise as determined by negative controls. Data were normalized by calculating the geometric mean of the top 100 miRNAs detected across all samples followed by quantile normalization.
Differential expression analysis
MiRNAs with an average expression level of at least 50 counts in at least one experimental group were considered for further analysis of the respective model system. Differential expression was calculated by the edgeR software package as implemented in Tibco Spotfire (TIBCO Software). MiRNAs greater than 3-fold differentially expressed between compared groups and a P-value adjusted for multiple testing less than 0.05 (Benjamini–Hochberg procedure) were considered to be significantly differentially expressed.
Quantitative Reverse Transcription Polymerase Chain Reaction of Fecal Samples
MiRNAs included in 30 ng of total RNA were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA) and Megaplex RT Primers, Human Pool A v2.1 (Applied Biosystems) as described by the manufacturer. Complementary DNA was pre-amplified using Megaplex PreAmp Primers, Human Pool A v2.1 (Applied Biosystems), and the TaqMan PreAmp Master Mix (Applied Biosystems) according to the instructions of the manufacturer. Reactions were performed in triplicates using TaqMan MicroRNA Assays (Applied Biosystems) miR-16-5p (assay ID: 000391), miR-142-5p (002248), miR-223-3p (002295), miR-128-3p (002216), miR-27a-3p (000408), miR-10b-5p (002218), miR-375 (000564), miR-10a-5p (000387), miR-200a-3p (000502), miR-192-5p (000491), and miR-320a (002277) and the TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR was run on a 7900HT Fast Real-Time PCR System (Applied Biosystems) according to the cycling parameters recommended by the manufacturer. NormFinder algorithm26 was applied to identify the endogenous control miRNA, miR-320a (stability value = 0.195), based on the small RNA-seq data of the fecal samples selected for quantitative reverse transcription–polymerase chain reaction (qRT-PCR) confirmation. In total 74 miRNAs with a mean expression of at least 10 log2 cpm were considered for NormFinder analysis. Threshold cycle (Ct) values greater than 35 were excluded from any calculations. Log2 fold changes (FCs) determined by qRT-PCR were calculated per CD patient and HC according to the 2−∆∆Ct method using mean HC expression as calibrator.
Correlation Analyses
Matching log2 FCs calculated for miRNAs of interest obtained from small RNA-seq and qRT-PCR analyses were compared using Pearson correlation. Correlation of log2 fecal miRNA levels with clinical CDAI and CDEIS scores was evaluated using Spearman rank correlation. Correlation analyses were calculated using GraphPad Prism (La Jolla, CA) and a significant correlation was defined by a P-value less than 0.05.
RESULTS
Fecal MiRNA Core Set
In order to investigate differential miRNA expression in patients with CD first a fecal miRNA core set was generated which contained fecal miRNAs that most likely originated from the intestinal tissue and were robustly expressed.
Quality assessment of normalized miRNA profiles obtained from 68 different intestinal biopsies by principal component analysis revealed 3 outliers with aberrant expression profiles that were excluded for further analyses. Normalized miRNA read count data of the remaining 65 samples consisting of 32 ileal and 33 colonic biopsies were used to define the biopsy miRNA set. This set comprised miRNAs with an expression level of at least 2 log2 cpm in at least 16 samples (corresponds to 50% of ileal sample size). The principal component analysis revealed that the individual ileal and colonic expression profiles were equally distributed (Supplementary Figure S1). Taken together, filtering of intestinal small RNA-seq data resulted in 456 abundantly expressed miRNAs (Supplementary Figure S2A).
Consistent with the filtering of the intestinal RNA-seq data, normalized fecal small RNA-seq data were filtered for miRNAs with expression levels of at least 2 log2 cpm in more than 8 samples (corresponds to 50% of HC sample size) which represented the preliminary fecal miRNA set of 215 miRNA species (Supplementary Figure S2B). Due to the high variability in the detection of low-expressed miRNAs across the fecal samples, we removed samples for further analyses if less than 50% of the preliminary fecal miRNA set was detected. This quality control filter necessitated the exclusion of 13 fecal CD samples. Then, the preliminary fecal miRNA set was compared to the biopsy miRNA set in order to identify fecal miRNAs which potentially originated from the intestinal tissue. Fecal miRNAs that emerged in both sets defined the fecal miRNA set which included 190 miRNAs (Supplementary Figure S2A and B). To evaluate robustly expressed miRNAs the fecal miRNA set was restricted to miRNAs with levels of at least 10 log2 cpm in at least 50% of CD or HC samples, respectively, which was further defined as the fecal miRNA core set of 89 miRNAs (Supplementary Figure S2B).
Differential Fecal MiRNA Expression
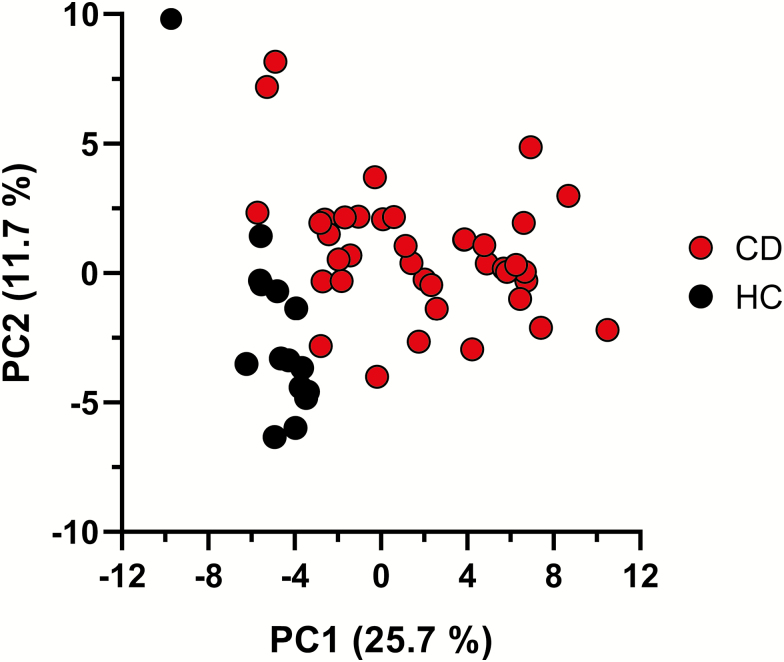
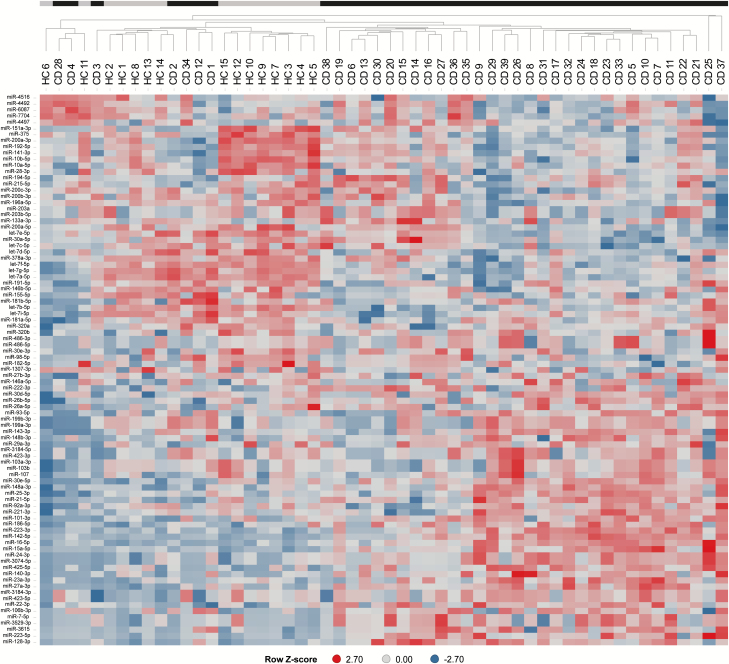
Principal component analysis (Fig. 1) revealed that the fecal miRNA core set expression profiles separated the patients with CD from HC. Fecal miRNA expression profiles from patients with CD were characterized by a higher variability compared to those of HC. Separation of the patients with CD from the HC was further substantiated by hierarchical clustering analysis (Fig. 2).
Figure 1.
Principal component analysis of fecal miRNA expression profiles. Displayed are the first 2 components from the principal component analysis of the 39 patients with CD and 15 HC. Expression data of the 89 robustly detected fecal miRNAs were included.
Figure 2.
Hierarchical clustering of CD patients and healthy controls based on fecal miRNA expression profiles. Black bars indicate patients with CD (CD1–CD39) and gray bars indicate healthy controls (HC1–HC15). Scaled miRNA expression levels are shown as Z-scores of the log2-transformed counts per million miRNA read counts.
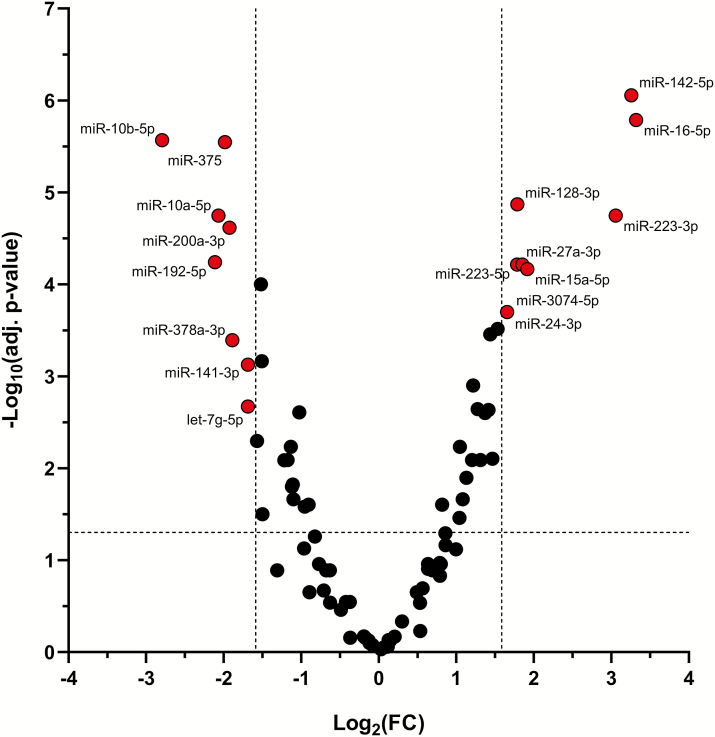
Differential miRNA expression analysis revealed that 17 miRNAs were significantly deregulated more than 3-fold. The expression levels of the 9 miRNAs miR-15a-5p, miR-16-5p, miR-128-3p, miR-142-5p, miR-24-3p, miR-27a-3p, miR-223-3p, miR-223-5p, and miR-3074-5p were significantly increased, whereas the expression levels of the 8 miRNAs miR-10a-5p, miR-10b-5p, miR-141-3p, miR-192-5p, miR-200a-3p, miR-375, miR-378a-3p, and let-7g-5p were significantly decreased in fecal CD samples (Fig. 3, Table 2). The highest FC of miRNA levels was detected for miR-16-5p (FC = 10), miR-142-5p (FC = 9.6), and miR-223-3p (FC = 8.3). The lowest FC was observed for miR-10b-5p (FC = −6.9), miR-192-5p (FC = −4.3), and miR-10a-5p (FC = −4.2). The functions of the differentially expressed fecal miRNAs are summarized according to published data in Table 2. Fecal miRNAs that did not meet our stringent significance criteria but were more than 2-fold (adj. P < 0.05) deregulated are listed in Supplementary Table S1 and Supplementary Table S2.
Figure 3.
Differentially expressed fecal miRNAs in CD patients compared to healthy controls. The expression of 17 fecal miRNAs (displayed in red) was significantly [absolute fold change (FC) >3, adj. P < 0.05] altered in patients with CD (n = 39) compared to healthy controls (n = 15).
Table 2.
Fecal miRNAs Significantly Increased and Decreased in Patients with Crohn’s Disease
| MiRNA | Fold Change | Adj. P | Target | Reference (PubMed ID) |
|---|---|---|---|---|
| miR-16-5p | 10.0 | 1.6 × 10–6 | TNF-α, IL-12/IL-23p40, Rictor | 26538392, 26073885, 15766526 |
| miR-142-5p | 9.6 | 8.8 × 10–7 | — | — |
| miR-223-3p | 8.3 | 1.8 × 10–5 | C/EBPβ, STAT3, Roquin, Claudin-8, FOXO3a | 27029486, 26878986, 26526992, 22937006, 22043014 |
| miR-15a-5p | 3.8 | 6.8 × 10–5 | — | — |
| miR-27a-3p | 3.6 | 6.1 × 10–5 | Runx1 | 19298589 |
| miR-128-3p | 3.5 | 1.3 × 10–5 | M-CSF | 24415783 |
| miR-223-5p | 3.4 | 6.1 × 10–5 | — | — |
| miR-24-3p | 3.2 | 2.0 × 10–4 | — | — |
| miR-3074-5p | 3.2 | 2.0 × 10–4 | — | — |
| miR-141-3p | −3.2 | 7.5 × 10–4 | CXCL12β, CXCL2, TGF-β2 | 18835392, 20952520, 24000293 |
| let-7g-5p | −3.2 | 2.1 × 10–3 | THBS1, TGFBR1, SMAD2 | 24291274 |
| miR-378a-3p | −3.7 | 4.1 × 10–4 | — | — |
| miR-200a-3p | −3.8 | 2.4 × 10–5 | CXCL2, TGF-β2 | 18835392, 20952520 |
| miR-375 | −4.0 | 2.8 × 10–6 | JAK2 | 24718681 |
| miR-10a-5p | −4.2 | 1.8 × 10–5 | NOD2, IL-12/IL-23p40 | 25281418, 22068236 |
| miR-192-5p | −4.3 | 5.7 × 10–5 | NOD2, CXCL2 | 18835392, 24297055 |
| miR-10b-5p | −6.9 | 2.7 × 10–6 | — | — |
Fecal miRNAs greater than 3-fold increased or decreased (adj. P < 0.05) in patients with Crohn’s disease compared to healthy controls. Targets are experimentally confirmed as described in the references.
Expression of the 5 most significantly up-regulated miRNAs miR-16-5p, miR-142-5p, miR-223-3p, miR-15a-5p, and miR-27a-3p as well as the 5 most significantly down-regulated miRNAs miR-10b-5p, miR-192-5p, miR-10a-5p, miR-375, and miR-200a-3p were confirmed for a subset of 10 CD patients and 5 HC using qRT-PCR. Correlation analysis revealed that the small RNA-seq data significantly correlated with the qRT-PCR data (R2 = 0.76, P < 0.001, Supplementary Figure S3).
Correlation of Fecal MiRNA Levels with Clinical Scores
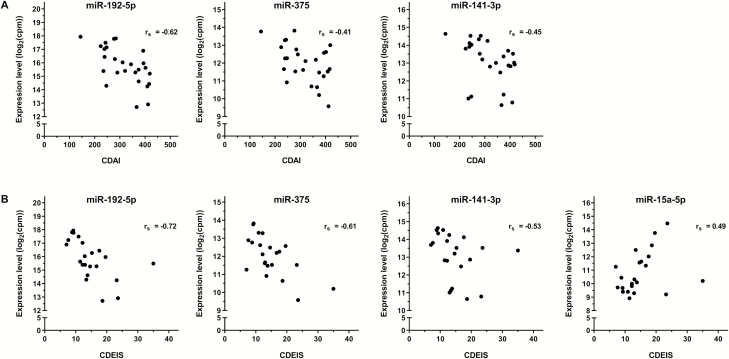
In order to investigate whether differences in fecal miRNA expression might reflect the degree of disease activity, fecal miRNA levels were correlated with the CDAI and CDEIS scores. Correlation analysis revealed a significantly negative correlation of the CDAI and CDEIS with miR-192-5p (rs_CDAI = −0.62; rs_CDEIS = −0.72), miR-375 (rs_CDAI = −0.41; rs_CDEIS = −0.61) and miR-141-3p (rs_CDAI = −0.45; rs_CDEIS = −0.53) expression levels (Fig. 4). Moreover, the expression of miR-15a-5p positively correlated with the CDEIS (rs_CDEIS = 0.49, Fig. 4). Correlation scores of all significantly deregulated fecal miRNAs are summarized in Supplementary Table S3.
Figure 4.
Correlation of fecal miRNA expression with CDAI and CDEIS. Matched fecal miRNA expression [log2 counts per million miRNA read counts (cpm)] and Crohn’s disease activity index (CDAI, n = 26) scores showed a significantly (P < 0.05) inverse correlation for miR-192-5p, miR-375, and miR-141-3p (A). Fecal levels of the same miRNAs showed also a significant inverse correlation with Crohn’s disease endoscopic index of severity (CDEIS, n = 22) scores. A significantly positive correlation with the CDEIS was observed for miR-15a-5p (B).
Back-Translation into Animal Models
In addition to human samples, fecal miRNA profiles were investigated in mouse models of DSS, T-cell transfer, and H. typhlonius/stress-induced intestinal inflammation. Differential expression analysis identified 72 (43 up, 29 down), 70 (41 up, 29 down), and 12 (10 up, 2 down) significantly more than 3-fold differentially expressed miRNAs at day 6, 13, and 21 of DSS-induced inflammation, respectively. For the adoptive T-cell transfer model 12 (11 up, 1 down) miRNAs were significantly differentially expressed whereas 5 (1 up, 4 down) significantly differentially expressed miRNAs were identified for the stress model.
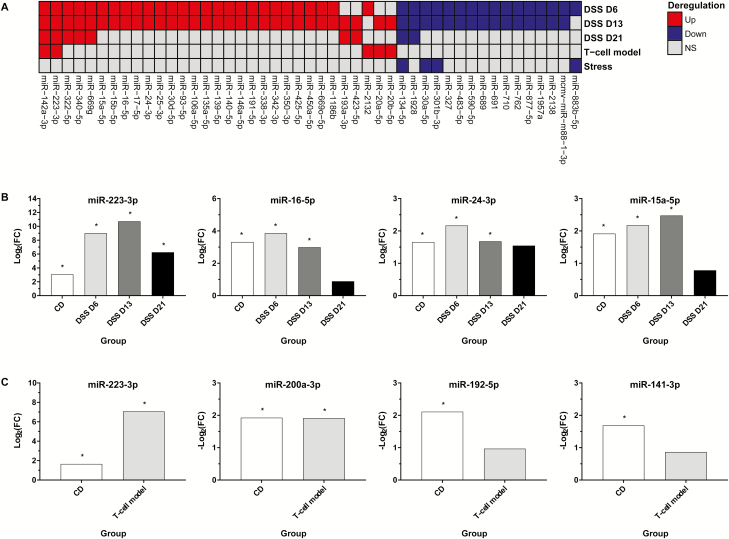
A comparison of the differentially expressed miRNA sets showed that many up-regulated miRNAs were shared for the different time points of DSS-induced inflammation. For day 6 and 13, 26 commonly up-regulated fecal miRNAs were identified. Five of the 26 miRNAs were also found on day 21 including miR-142a-3p, miR-223-3p, miR-322-5p, miR-340-5p, and miR-669g (Fig. 5A). Additionally, miR-223-3p and miR-142a-3p were up-regulated in the T-cell transfer model which in total shared 5 miRNAs with the DSS model (Fig. 5A). The stress model, in contrast, did not share any up-regulated miRNA with the remaining models while the 4 down-regulated miRNAs were also found in the DSS model (Fig. 5A). One of those miRNAs, miR-134-5p, was significantly decreased in all model systems except the T-cell transfer model. Similar to the up-regulated miRNAs most down-regulated miRNAs were shared by the DSS model on days 6 and 13 represented by 15 commonly decreased miRNAs (Fig. 5A).
Figure 5.
Comparison of fecal miRNA profiles in murine colitis models and human CD. Comparison of significantly up- (Up) and down-regulated (Down) fecal miRNAs on day 6 (D6), day 13 (D13), and day 21 (D21) of dextran sodium sulfate (DSS)-induced intestinal inflammation, the T-cell transfer model (T-cell model), and Helicobacter typhlonius/stress model (Stress). Fecal miRNAs that were significantly [absolute fold change (FC) >3, adj. P < 0.05; NS, not significant] deregulated in at least 2 murine models or time points of the DSS model were considered. MiR-17-5p and miR-106a-5p as well as miR-20a-5p and miR-20b-5p were targeted by the same probe, respectively (A). Several of the fecal miRNAs that were significantly (*) altered in patients with CD were also deregulated in the same direction in the DSS (B) and the T-cell transfer models (C).
Comparison of the 3 model systems to human CD revealed that several of the miRNAs that were significantly deregulated in human CD patients were also deregulated in the DSS model and the T-cell transfer model but not in the stress model. For the DSS model, this set of fecal miRNAs included miR-223-3p, miR-16-5p, miR-15a-5p, and miR-24-3p that were significantly increased on day 6 and 13 post-DSS treatment. On day 21 miR-223-3p was still significantly increased whereas miR-15a-5p, miR-16-5p, and miR-24-3p were not significantly altered but 1.7-fold to 2.9-fold increased (Fig. 5B). In line with the DSS model, a significantly increased miR-223-3p expression was detected for the T-cell transfer model whereas miR-200a-3p was 3.8-fold decreased which is also consistent with human CD. Decreased levels of miR-192-5p and miR-141-3p, as observed in human CD, were found in the T-cell transfer model; however, the FCs of 2.0 and 1.8 did not meet our stringent cutoff criteria (Fig. 5C).
DISCUSSION
This study provides evidence that the miRNA profile is altered in feces of patients with CD compared to HC, as well as in relevant animal model systems. Intestinal and fecal miRNA expression profiling provides a high overlap of robustly expressed miRNAs between both sample matrices. Out of 215 miRNAs detected in feces of both CD and HC the expression of 190 miRNAs was also abundant in intestinal biopsies. To further substantiate that the analyzed fecal miRNAs originate from intestinal tissue only miRNAs were evaluated which were strongly expressed in tissue biopsies resulting in a fecal miRNA core set of 89 miRNAs. Differential miRNA expression analysis identified 17 strongly (FC >3) deregulated miRNAs in patients with CD. Nine miRNAs were significantly increased whereas the levels of 8 miRNAs were significantly decreased in feces of patients with CD compared to HC. Based on published data there is strong evidence that most of the deregulated fecal miRNA species are implicated in hallmark pathways of CD, such as Th17 inflammation, intestinal fibrosis, epithelial barrier integrity, autophagy and cell proliferation, and/or differentiation processes.
CD is characterized by enhanced recruitment and retention of immune cells, such as macrophages, neutrophils, and T cells into the inflamed intestine, where they are activated and release pro-inflammatory cytokines. Previous studies reported that the decreased expression of miR-141-3p, miR-200a-3p, and miR-192-5p in inflamed intestinal tissues of patients with CD correlated with increased recruitment of immune cells.27,28 Notably miR-141 and its closely related family member miR-200a share the same 3′UTR seed sequence and modulate the expression of C-X-C motif chemokine (CXCL)12β, which is involved in promoting immune cell infiltrations.28,29 In colonic epithelial cells induction of CXCL2 expression by tumor necrosis factor alpha (TNFα) was accompanied by a concomitant reduction in miR-192. Our study revealed decreased fecal levels of the 2 miR-200 family members miR-141-3p and miR-200a-3p as well as miR-192-5p in patients with CD. The immune response of CD is dominated by Th1- and Th17-mediated inflammation. Two miRNAs, namely, miR-223 and miR-10a, are directly linked to both Th1- and Th17-mediated inflammation. Th1 and Th17 immune responses can be modulated by miR-10a via inhibiting IL-12/IL-23p40 expression.9 In healthy colon tissue, the balanced level of miR-223-3p and its target claudin-8 is responsible for the homeostasis of the intestinal barrier.13 Disturbed Th17 inflammation and intestinal barrier integrity are characterized by induced expression of miR-223-3p and reduced miR-10a levels in the intestinal tissue from patients with CD that were also depicted in feces of patients with CD compared to HC in our study.9,13 Increased levels of miR-223-3p were previously also detected in feces of patients with CD.23–25 The highest deregulated fecal miRNA in CD, miR-16-5p (FC >10), was also observed to be over-expressed in peripheral blood, serum, and feces of patients with inflammatory bowel disease.16,23,30 Aberrant nuclear factor-kappa B (NF-κB) signaling is a key driver of pro-inflammatory cytokines, such as TNFα, IL-6, and IL-8 during disease progression of CD.31 In particular, miR-16 negatively regulates the expression of the A2A adenosine receptor to control the activation of the NF-κB signaling pathway. Furthermore, translocation of the NF-κB p65 protein into the nucleus and expression of the pro-inflammatory cytokines interferon-γ and IL-8 is promoted by transfection of miR-16 mimics in colonic epithelial cells.30
Chronic inflammation is a precursor of intestinal fibrosis and epithelial to mesenchymal transition (EMT) in which TGF-β plays a major role. Members of the miR-200 family, such as miR-141-3p and miR-200a-3p, play a key role in EMT and TGF-β is a prominent target.32 TGF-β is strongly expressed in fibrotic colonic tissue whereas miR-141-3p and miR-200a-3p are decreased during fibrotic processes thus facilitating TGF-β-dependent EMT.29 Our study identified decreased levels of miR-141-3p and miR-200a-3p in patients with CD, which reflect the activated TGF-β signaling pathway during intestinal fibrotic processes.
Chronic inflammation promotes CRC development and long-standing CD is associated with an increased risk of CRC.33 Therefore, it is not surprising that miRNAs that are involved in cell differentiation and proliferation were deregulated in feces of patients with CD in our study. Examples are miRNAs that function as a tumor suppressor, such as miR-375,34 or have dual functions in CRC such as miR-15a and miR-16-5p.35
Our fecal miRNA core set that is characterized by high expression levels (>10 log2 cpm) was aimed to minimize unspecific miRNA sources, such as pancreatic juice. Altered fecal miRNA levels might derive from cell exfoliation resulting from increased shedding of colonocytes during perturbed intestinal homeostasis.36 MiRNAs can also be actively secreted from epithelial cells that line the colonic lumen or leak from inflamed and/or injured tissue. Cell-specific deletion of miRNAs in intestinal epithelial cells (IECs) using DicerΔIEC mice and in homeodomain-only protein (Hopx)-expressing cells, such as Paneth and goblet cells, using DicerΔHopx mice resulted in decreased fecal miRNA levels suggesting that IECs and Hopx-expressing cells are the major source of fecal miRNAs.37 This strongly supports our approach to investigate fecal miRNAs that were also detected in the intestinal tissue as potential CD biomarker candidates.
Recently, Wang et al.38 identified significantly increased levels of miR-223-3p in the serum of patients with CD compared to HC. Moreover, these levels correlated with disease activity scores, such as CDAI and simple endoscopic score for CD but with low Spearman correlation coefficients of 0.48 and 0.35, respectively. Here, we demonstrated that fecal levels of miR-192-5p, miR-375, and miR-141-3p correlated with the CDAI and miR-192-5p, miR-375, miR-141-3p, and miR-15a with the CDEIS with Spearman correlation coefficients of rs_CDEIS = −0.72 as observed for miR-192-5p. The miR-192-5p–CDEIS correlation is comparable with those of the most commonly used fecal biomarkers, calprotectin and lactoferrin.39 Interestingly, fecal miRNAs might also serve as promising treatment effect markers. The expression of miR-10a-5p, a miRNA that was significantly decreased in fecal samples of patients with CD in our study, was previously shown to be decreased in the inflamed intestinal mucosa of patients with CD compared to HC. Anti-TNFα therapy with infliximab restored miR-10a expression levels in the intestinal mucosa of active patients with CD.9 In a recent study we showed that miR-223-3p levels decreased significantly in colon biopsies and feces of patients with CD treated with risankizumab, which further substantiates the potential of fecal miRNAs as treatment effect markers.40
In our translational study, we also show that fecal miRNA profiles differentiate diseased from control mice of the DSS, adoptive T-cell transfer, and H. typhlonius/stress-induced colitis models. Moreover, consistent with human CD, significantly increased fecal levels of miR-223-3p, miR-16-5p, miR-15a-5p, and miR-24-3p and decreased levels of miR-200a-3p were observed for the DSS and/or the T-cell transfer model. In line with observations in human CD, increased intestinal tissue levels of miR-223-3p were previously also reported for DSS-treated mice.11,12 Most of these miRNAs are linked to key CD pathways. Our back-translational approach therefore confirms that CD-associated alterations of the human fecal miRNA profile are reflected in the preclinical mouse models, although to a different degree. The patients with CD included in this study suffered from mild to severe CD, whereas the H. typhlonius/stress-induced colitis model represents a rather mild model that may explain the difference of the deregulated fecal miRNA sets. Our study can therefore also support the selection of a relevant murine model system to investigate certain miRNA-linked and CD-associated pathways. Differential expression of the CD-linked fecal miRNAs miR-16-5p and miR-15a-5p along with the different time points of the DSS model furthermore suggests that fecal miRNAs may be useful biomarkers to assess disease progression. In humans, further longitudinal studies are needed to confirm the fecal miRNA profiles observed in this cross-sectional study and to evaluate whether fecal miRNAs can provide more precise insights into disease progression. These studies should therefore also compare the inflammatory CD phenotype, which was investigated in our study, with different CD phenotypes such as stenotic CD to evaluate whether fecal miRNAs can distinguish the different CD phenotypes.
CONCLUSIONS
The present work provides first insight into altered fecal miRNA profiles in patients with CD and relevant mouse models. These novel, noninvasive markers show promise as tools for the mechanistic investigation of intestinal pathophysiology, including disease progression, and display potential to monitor treatment effects.
Supplementary Material
ACKNOWLEDGMENTS
We thank Germán G. Leparc for sharing his small RNA-seq analysis workflow.
Funding: This work was supported by Boehringer Ingelheim Pharma GmbH & Co. KG.
Conflict of Interest: All authors except S.O.R. and D.L. are employees of Boehringer Ingelheim. S.O.R. and D.L. have no conflict of interest to disclose.
Author Contributions: P.B., D.D., F.G., I.H., W.O.B., and S.V. contributed to the conception and design of the human CD study. C.T.W., E.K., K.R., M.R., I.J., C.E., and R.V. contributed to data acquisition of the human CD study and DSS model as well as data analysis. S.O.R. and D.L. contributed to the conception, design, data acquisition, and analysis of the social stress model. F.S., M.P., and E.R. contributed to the conception, design, data acquisition, and analysis of the T-cell transfer model. C.T.W., P.B., D.D., R.S., B.S., F.W., D.L., and S.O.R. contributed to data analysis and data interpretation. P.B., D.D., and C.T.W. wrote the manuscript. All authors contributed to scientific discussions, revision of the manuscript, and had full access to all the data and agreed to submit for publication.
REFERENCES
- 1. Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380:1590–1605. [DOI] [PubMed] [Google Scholar]
- 2. Loddo I, Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol. 2015;6:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sands BE. Biomarkers of inflammation in inflammatory bowel disease. Gastroenterology. 2015;149:1275–1285.e2. [DOI] [PubMed] [Google Scholar]
- 5. Kalla R, Ventham NT, Kennedy NA, et al. . MicroRNAs: new players in IBD. Gut. 2015;64:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fasseu M, Treton X, Guichard C, et al. . Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:e13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu F, Zhang S, Dassopoulos T, et al. . Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16:1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brest P, Lapaquette P, Souidi M, et al. . A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–245. [DOI] [PubMed] [Google Scholar]
- 9. Wu W, He C, Liu C, et al. . miR-10a inhibits dendritic cell activation and Th1/Th17 cell immune responses in IBD. Gut. 2015;64:1755–1764. [DOI] [PubMed] [Google Scholar]
- 10. Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. J Immunol. 2011;187:5834–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou H, Xiao J, Wu N, et al. . MicroRNA-223 Regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPβ. Cell Rep. 2015;13:1149–1160. [DOI] [PubMed] [Google Scholar]
- 12. Neudecker V, Haneklaus M, Jensen O, et al. . Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H, Chao K, Ng SC, et al. . Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Philpott DJ, Sorbara MT, Robertson SJ, et al. . NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. [DOI] [PubMed] [Google Scholar]
- 15. Guo Z, Wu R, Gong J, et al. . Altered microRNA expression in inflamed and non-inflamed terminal ileal mucosa of adult patients with active Crohn’s disease. J Gastroenterol Hepatol. 2015;30:109–116. [DOI] [PubMed] [Google Scholar]
- 16. Paraskevi A, Theodoropoulos G, Papaconstantinou I, et al. . Circulating microRNA in inflammatory bowel disease. J Crohn’s Colitis. 2012;6:900–904. [DOI] [PubMed] [Google Scholar]
- 17. Iborra M, Bernuzzi F, Correale C, et al. . Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol. 2013;173:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu F, Guo NJ, Tian H, et al. . Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollis M, Nair K, Vyas A, et al. . MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Link A, Balaguer F, Shen Y, et al. . Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Link A, Becker V, Goel A, et al. . Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS One. 2012;7:e42933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ahmed FE, Ahmed NC, Vos PW, et al. . Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. proof of principle. Cancer Genomics Proteomics. 2013;10:93–113. [PubMed] [Google Scholar]
- 23. Schönauen K, Le N, von Arnim U, et al. . Circulating and fecal microRNAs as biomarkers for inflammatory bowel diseases. Inflamm Bowel Dis. 2018;24:1547–1557. [DOI] [PubMed] [Google Scholar]
- 24. Ji Y, Li X, Zhu Y, et al. . Faecal microRNA as a biomarker of the activity and prognosis of inflammatory bowel diseases. Biochem Biophys Res Commun. 2018;503:2443–2450. [DOI] [PubMed] [Google Scholar]
- 25. Verdier J, Breunig IR, Ohse MC, et al. . Faecal micro-RNAs in inflammatory bowel diseases. J Crohn’s Colitis. 2020;14:110–117. [DOI] [PubMed] [Google Scholar]
- 26. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. [DOI] [PubMed] [Google Scholar]
- 27. Wu F, Zikusoka M, Trindade A, et al. . MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635.e24. [DOI] [PubMed] [Google Scholar]
- 28. Huang Z, Shi T, Zhou Q, et al. . miR-141 regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn’s disease. Gut. 2014;63:1247–1257. [DOI] [PubMed] [Google Scholar]
- 29. Wang B, Koh P, Winbanks C, et al. . miR-200a prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes. 2011;60:280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tian T, Zhou Y, Feng X, et al. . MicroRNA-16 is putatively involved in the NF-κB pathway regulation in ulcerative colitis through adenosine A2a receptor (A2aAR) mRNA targeting. Sci Rep. 2016;6:30824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y, Ge W, Xu L, et al. . miR-200b is involved in intestinal fibrosis of Crohn’s disease. Int J Mol Med. 2012;29:601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. [DOI] [PubMed] [Google Scholar]
- 34. Mao Q, Quan T, Luo B, et al. . MiR-375 targets KLF4 and impacts the proliferation of colorectal carcinoma. Tumour Biol. 2016;37:463–471. [DOI] [PubMed] [Google Scholar]
- 35. Huang E, Liu R, Chu Y. miRNA-15a/16: as tumor suppressors and more. Future Oncol. 2015;11:2351–2363. [DOI] [PubMed] [Google Scholar]
- 36. Vincent C, Mehrotra S, Loo VG, et al. . Excretion of host DNA in feces is associated with risk of Clostridium difficile infection. J Immunol Res. 2015;2015:246203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu S, da Cunha AP, Rezende RM, et al. . The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H, Zhang S, Yu Q, et al. . Circulating microRNA223 is a new biomarker for inflammatory bowel disease. Medicine (Baltimore). 2016;95:e2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sipponen T, Savilahti E, Kolho KL, et al. . Crohn’s disease activity assessed by fecal calprotectin and lactoferrin: correlation with Crohn’s disease activity index and endoscopic findings. Inflamm Bowel Dis. 2008;14:40–46. [DOI] [PubMed] [Google Scholar]
- 40. Visvanathan S, Baum P, Salas A, et al. . Selective IL-23 inhibition by risankizumab modulates the molecular profile in the colon and ileum of patients with active Crohn’s disease: results from a randomised phase II biopsy sub-study. J Crohn’s Colitis. 2018;12:1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.