Abstract
Aim
Low-temperature plasma (LTP) has potential applications in cancer therapy. Herein, we explored the molecular mechanisms of proliferation inhibition induced by LTP.
Methods
LTP was generated by a helium atmospheric-pressure plasma jet and used to treat A549 and H1299 cells. CCK-8 and cell apoptosis assays were performed to evaluate the effects of LTP treatment on A549 and H1299 cells. The qRT-PCR was performed to measure the expression of miR-203a after treating with LTP. CCK-8, colony formation, cell apoptosis assays, and Western blotting were performed to analyse the function of miR-203a in the development of lung cancer. Dual-luciferase assay and Western blotting were used to probe the relationship between miR-203a and BIRC5, and gene silencing using si-BIRC5 was carried out to explore the effect of knocking down BIRC5 on lung cancer cells.
Results
We found that LTP significantly suppressed proliferation and promoted apoptosis in A549 and H1299 cells. The miR-203a expression was increased after cells were treated with LTP. The miR-203a expression was downregulated among lung cancer tissue samples, and overexpression of miR-203a suppressed cell growth and induced apoptosis in lung cancer cells. We showed that miR-203a targeted BIRC5. Moreover, silencing of BIRC5 caused proliferation inhibition and induced apoptosis in lung cancer cells.
Conclusion
Our study revealed that LTP inhibited proliferation and induced apoptosis in A549 and H1299 cells through the miR-203a/BIRC5 axis. These findings showed that LTP could potentially be used to treat lung cancer.
Keywords: low-temperature plasma, lung cancer, proliferation, miR-203a, BIRC5
Introduction
Lung cancer is the leading cause of cancer-related deaths among men worldwide. Currently, treatment of lung cancer includes surgery, chemotherapy, targeted therapy, radiation, radiofrequency ablation, or a combination of these therapies.1,2 However, treatment outcomes are generally poor. Therefore, to explore effective therapeutic strategies for lung cancer is essential. Low-temperature plasma (LTP) is a useful tool with several applications in the biomedical field, such as wound healing,3 teeth treatment,4 blood coagulation5 and tumour therapy. Recent studies showed that LTP is a promising alternative treatment for cancer.
MicroRNAs (miRNAs) can regulate gene expression by targeting mRNAs, and function as tumour suppressors or oncogenes in cancer cells.6 The miRNAs regulate a variety of complex biological processes including apoptosis and cell growth, proliferation and differentiation. The miRNA dysfunction is associated with cancer pathogenesis and development. Dysfunction of miR-101 is involved in ovarian cancer,7 miR-647 is related to gastric cancer,8 miR-130a is associated with colorectal cancer,9 miR-218 is linked to lung cancer,10 and miR-320a mediates liver cancer.11 The miR-203, located on chromosome 14, is involved in various types of cancer. miR-203a-3p functions as a tumour suppressor in gastric cancer,12 and miR-203a inhibits HCC cell invasion and metastasis.13
miR-203a (also known as a member of miR-203) may play a role as a tumour suppressor gene. Herein, we found that LTP could suppress the growth of lung cancer cells. We analysed the biological mechanisms of LTP in proliferation inhibition and apoptosis induction in lung cancer cells at the cellular and molecular levels. We also identified miRNAs altered after LTP treatment. We found that LTP mediated the expression of miR-203a, which targeted BIRC5 to inhibit proliferation and induce apoptosis. These findings demonstrated that LTP may have therapeutic potential for lung cancer.
Materials and Methods
Cell Culture and Tissues
A549, H1299, and HEK293 cells were cultured in DMEM with 10% fetal bovine serum (Gibco BRL, NY, USA) in a humidified incubator with 5% CO2 at 37°C. Cell line were provided by the Key University, Ministry of Education (Xi’an, Shaanxi, China). The use of these cell lines was approved by Institute Research Ethics Committee at Cancer, Xi’an Jiaotong University. Human lung cancer tissues and the matched adjacent non-malignant tissues were obtained from the First Affiliated Hospital of Xi’an Jiaotong University. The patient consent was written informed consent. The study was approved by the Institute Research Ethics Committee at Cancer, Xi’an Jiaotong University.
LTP Treatment Procedure
A549 and H1299 cells were cultured in 96-well plates in 200 μL of medium per well at a density of 2.5 × 104 cells/mL in a humidified incubator with 5% CO2 overnight. The cells were then treated with atmospheric helium plasma at room temperature for 0, 5, 10, 15, 20, 25, or 30 s and cultured for 24, 48, or 72 h.
qRT-PCR
RNA was extracted by TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Prime-Script RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) and RNA were used to generate cDNA. cDNA was subjected to qRT-PCR using the SYBR Green PCR kit (Takara Biotechnology Co., Ltd.). An FTC-3000™ system (Funglyn Biotech Inc., Toronto, ON, Canada) was used to perform PCR amplification. The primer sequences are presented in Table 1.
Table 1.
Primers and oligonucleotides used in this work
| Name | Sequence (5′–3′) |
|---|---|
| miR-203a-RT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTAGTGG |
| miR-203a-F | ATCCAGTGCGTGTCGTG |
| miR-203a-R | TGCTGTGAAATGTTTAGGA |
| U6-RT | CGCTTCACGAATTTGCGTGTCAT |
| U6-F | GCTTCGGCAGCACATATACTAAAAT |
| U6-R | CGCTTCACGAATTTGCGTGTCAT |
| miR-203a inhibitor | CTAGTGGTCCTAAACATTTCAC |
| inhibitor-ctrl | CAGTACTTTTGTGTAGTACAA |
| si-ctrl-S | UUCUCCGAACGUGUCACGUTT |
| si-ctrl-A | ACGUGACACGUUCGGAGAATT |
| si-BIRC5-S | CCAACAAUAAGAAGAAAGATT |
| si-BIRC5-A | UCUUUCUUCUUAUUGUUGGTT |
| BIRC5-F | CTTGGCCCAGTGTTTCTTCT |
| BIRC5-R | CTTATTGTTGGTTTCCTTTGCAT |
| β-Actin-F | CCAACCGCGAGAAGATGA |
| β-Actin-R | CCAGAGGCGTACAGGGATAG |
Plasmid Construction and Transient Transfection
Pre-miR-203a oligonucleotides were synthesized and cloned into the pcDNA6.2-GW/EmGFP vector between the EcoRI and HindIII sites. We identified a fragment of BIRC5 as an miR-203a target through bioinformatic analysis. The 3′ UTR of BIRC5 was synthesized and cloned into the pmirGLODual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) between the SacI and XhoI sites. The sequences of miR-203a inhibitor and inhibitor-control are listed in Table 1. Small interfering RNA (siRNA) targeting BIRC5 were purchased from GenePharma (GenePharma, Shanghai, China). All sequences are listed in Table 1. A549 and H1299 cells were seeded in DMEM supplemented with 10% FBS and cultured for 24 h. Then, the miR-203a overexpression vector, miR-ctrl, miR-203a inhibitor, inhibitor-ctrl, si-BIRC5, or si-ctrl was transfected into the cells by jetPRIME® from Polyplus-transfection (Illkirch, France).
CCK-8 Assay
A549 and H1299 cells were seeded in 96-well plates. Cells were treated by LTP or transfected with miR-203a, miR-203a inhibitor, si-BIRC5, or their respective controls. Cell viability was analyzed by the Cell Counting Kit-8 (CCK-8, 7Sea Biotech, Shanghai, China) at 24, 48, and 72 h after LTP treatment or transfection. The optical density at a wavelength of 450 nm was measured using the FLUOstar OPTIMA microplate reader (BMG Labtech GmbH, Ortenberg, Germany).
Colony Formation Assay
A549 and H1299 cells (3000 cells/well) were seeded into 6-well plates after transfection. The plates were washed with phosphate-buffered saline (PBS) after incubation for 14 days, and the colonies were stained with 0.1% crystal violet for 30 min. The colonies were then rinsed with phosphate-buffered saline and analyzed using a computer software (Quantity One, Bio-Rad, Hercules, CA, USA).
Cell Apoptosis Assay
A549 and H1299 cells treated with LTP or transfection were harvested after 48 h of culture and stained with the annexin V-FITC/PI Apoptosis Detection Kit (7Sea Biotech, Shanghai, China). The apoptosis of cells was examined by flow cytometry (Becton Dickinson, USA).
Immunohistochemistry
The tissues were fixed in 10% formalin buffer and made into paraffin sections. The paraffin sections were sliced into a thickness of 5 µm. The sections were deparaffinized with xylene and hydrated using graded alcohol, antigen retrieval and blocking. The slides were incubated with primary antibodies (BIRC5, diluted 1:100) at 4°C overnight, then the slides were incubated with secondary antibody for 30 min at room temperature. Detection was performed using 3, 3′-diaminobenzidine (DAB) and hematoxylin. Finally, digital images were obtained using a Leica image analysis system.
Dual-Luciferase Assay
The 3′ UTR of BIRC5 including wild-type (wt) or mutated (mut) miR-203a binding sites was cloned in the pmirGLO Vector (Promega, USA). HEK293 cells were co-transfected with pre-miR-203a and wt BIRC5-3′ UTR, mut BIRC5-3′ UTR, or pmirGLO vector using jetPRIME®. A reporter assay was employed via the dual-luciferase reporter assay system (Promega) at 24 h post-transfection.
Western Blot
Proteins were extracted with the radioimmunoprecipitation assay buffer. The proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% non-fat milk in Tris buffer saline with 0.1% Tween-20 (TBST) for 1 h. Then incubated with antibodies against BIRC5 (ProteinTech Group, Wuhan, China; diluted 1:1000) and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA; diluted 1:2000) at 4°C overnight. After three washes with TBST, the membrane was incubated with a goat anti-rabbit or goat anti-mouse antibody (Bioworld; diluted 1/3000) for 2 h. TBST was used to wash the membrane for three times, then the membrane was incubated with the ECL Plus Reagent (Millipore, USA). The blots were detected by Quantity One imaging software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical Analysis
All data are presented as mean ± standard error. Data were analyzed by Student’s t-test or one-way ANOVA followed by a Tukey’s post hoc test to analyze the differences between the groups. All analyses were conducted using SPSS 13.0. p < 0.05 was considered to be statistically significant.
Results
Atmospheric-Pressure Plasma Jet (APPJ) Equipment
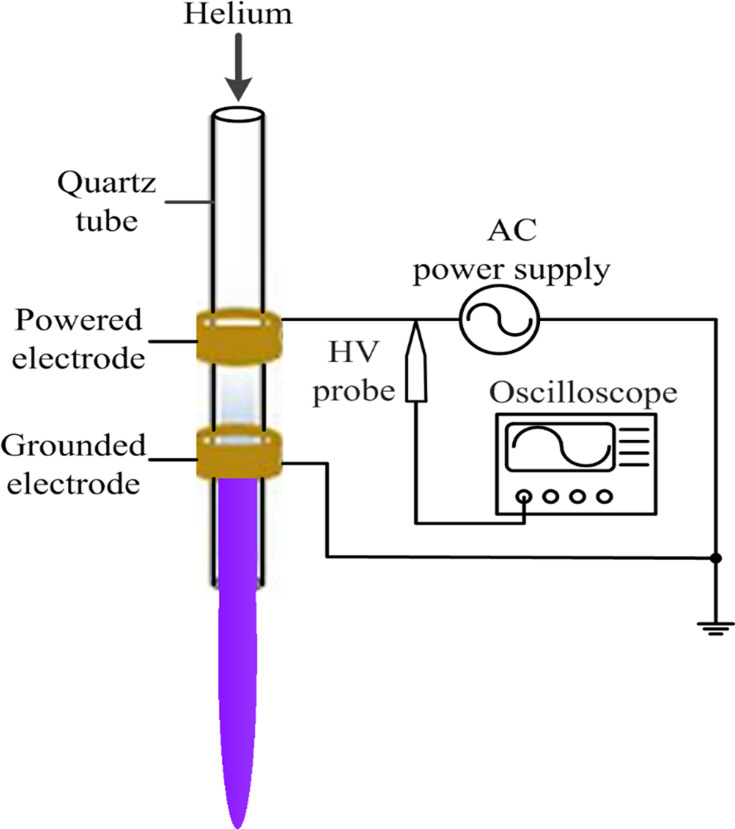
An APPJ with a co-axial double ring electrode configuration provided the plasma source. A hollow quartz tube with an inner diameter of 2 mm and an outer diameter of 4 mm was used as the barrier dielectric. The tube, which functioned as the barrier dielectric and airflow channel, was wrapped with two copper ring electrodes. The distance from the grounded electrode to the quartz tube nozzle was 0.5 cm. During the experiment, the helium flow rate was 1 L/min, the intermediate frequency was 39.5 kHz and the peak voltage was 7 kV. The length of the plasma jet generated by the APPJ was ~1.5 cm and the temperature at the tip of the effluent plasma ranged from 25°C to 34°C (Figure 1).
Figure 1.
Schematic of the experimental setup.
LTP Suppresses Lung Cancer Cell Proliferation and Induces Expression of miR-203a
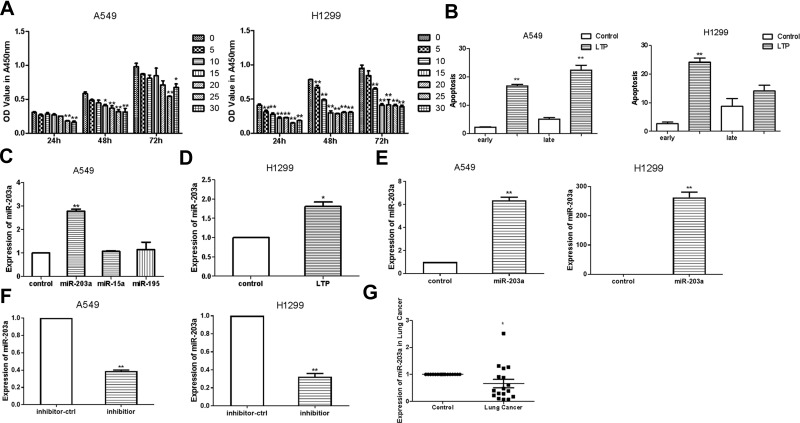
A549 and H1299 cells were seeded in 96-well plates and cultured for 24 h. They were then treated with atmospheric helium plasma at room temperature for various durations. CCK-8 assays indicated that LTP significantly inhibited the proliferation of lung cancer cells; when cells were treated for 25 s, growth inhibition almost reached a plateau compared to controls (Figure 2A). Therefore, we chose a treatment time of 25 s for further studies. Cell apoptosis assays showed that LTP induced apoptosis in A549 and H1299 cells (Figure 2B).
Figure 2.
LTP suppresses lung cancer cells proliferation and induces the expression of miR-203a. (A) Effects of LTP on A549 cells and H1299 cell proliferation were detected with CCK-8 assay. (B) Apoptosis was determined after A549 cells/H1299 cells were treated with LTP for 25 s. (C) qRT-PCR was performed to examine the expression of miR-203a, miR-15a and miR-195 after treating with LTP in A549 cells. (D) qRT-PCR was performed to examine the expression of miR-203a after treating with LTP in H1299 cells. (E) The expression of miR-203a was detected after transfected with pre-miR-203a comparing with miR-ctrl. (F) The expression of miR-203a was detected after transfected with inhibitor of miR-203a comparing with inhibitor-ctrl. (G) qRT-PCR was performed to examine miR-203a expression in17 paired lung cancer and adjacent normal tissues (*P<0.05, **P<0.01).
Next, we performed qRT-PCR to evaluate the expression of miR-203a, miR-15a and miR-195 following LTP treatment in A549 cells. Our results showed that miR-203a was upregulated after cells were treated with LTP (Figure 2C and D), hence we choose miR-203a for further study. We engineered an overexpression vector, synthesised an miR-203a inhibitor, and evaluated their effectiveness by qRT-PCR. The miR-203a overexpression vector significantly increased the expression of miR-203a, while the miR-203a inhibitor significantly suppressed the expression of miR-203a (Figure 2E and F). Expression of miR-203a was detected in 17 pairs of lung cancer tissues and their matched adjacent non-cancerous tissue samples obtained from patients at the First Affiliated Hospital of Xi’an Jiaotong University, China. Informed consent was obtained from each patient. We observed that miR-203a was significantly downregulated in lung cancer samples compared with matched adjacent non-cancerous tissues (Figure 2G). These results suggested that LTP could inhibit cell proliferation, induce apoptosis and expression of miR-203a in lung cancer cells.
The miR-203a Inhibits Proliferation and Induces Apoptosis in Lung Cancer Cells
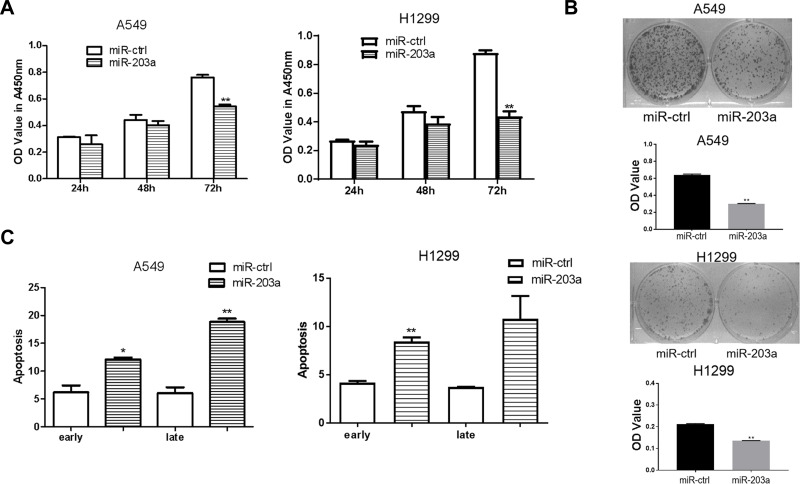
To confirm the effects of miR-203a on lung cancer cells, A549 and H1299 cells were transfected with pre-miR-203a or miR-ctrl. CCK-8 assays showed that overexpression of miR-203a inhibited cell proliferation (Figure 3A). Cells transfected with pre-miR-203a formed fewer colonies (Figure 3B). Cell apoptosis assays also showed that miR-203a overexpression induced a remarkable increase in the number of early apoptotic cells (Figure 3C).
Figure 3.
miR-203a inhibits proliferation and induces apoptosis in lung cancer cells. (A) A549/H1299 cells were transfected with pre-miR-203a or miR-ctrl, then detected with CCK-8 assay. (B) Colony formation was applied to measure the effect on A549/H1299 cells treated with miR-203a or miR-ctrl. (C) Apoptosis was detected in A549/H1299 cells after transfection with miR-203a or miR-ctrl (*P<0.05, **P<0.01).
The miR-203a Inhibitors Promote Proliferation in Lung Cancer Cells
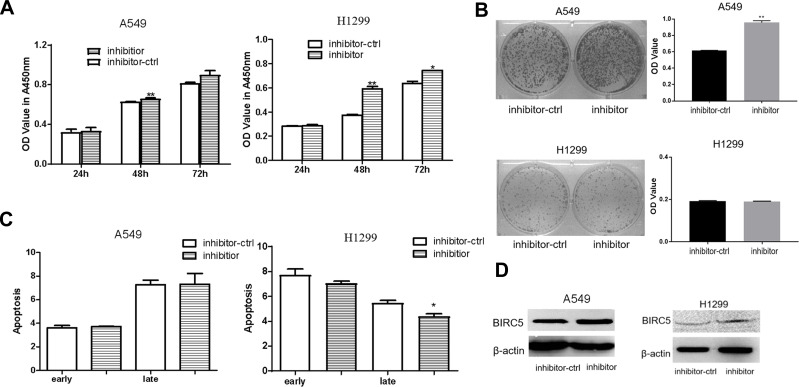
Silencing the expression of miR-203a increased the proliferation of lung cancer cells (Figure 4A), and the number of colonies formed was more in the miR-203a inhibitor-transfected than in the control in A549 cells(Figure 4B). There were no obvious differences between miR-203a inhibitor transfection and control inhibitor (inhibitor-ctrl) transfection in A549 cells, whereas the miR-203a inhibitor suppressed late apoptosis in H1299 cells (Figure 4C). BIRC5 protein expression was increased after miR-203a inhibitor transfection (Figure 4D).
Figure 4.
miR-203a inhibitor promotes proliferation in lung cancer cells. (A) miR-203a inhibitor or inhibitor-ctrl was transfection in A549/H1299 cells and determined by CCK-8 assay. (B) Colony formation was performed to detect the effect on A549/H1299 cells treated with miR-203a inhibitor or inhibitor-ctrl (*P<0.05, **P<0.01). (C) Apoptosis was detected in A549/H1299 cells after transfection with miR-203a inhibitor or inhibitor-ctrl (*P<0.05, **P<0.01). (D) BIRC5 protein expression levels were measured in A549/H1299 cells after transfection with miR-203a inhibitor or inhibitor-ctrl by Western blot (*P<0.05, **P<0.01).
BIRC5 Is a Direct Target of miR-203a
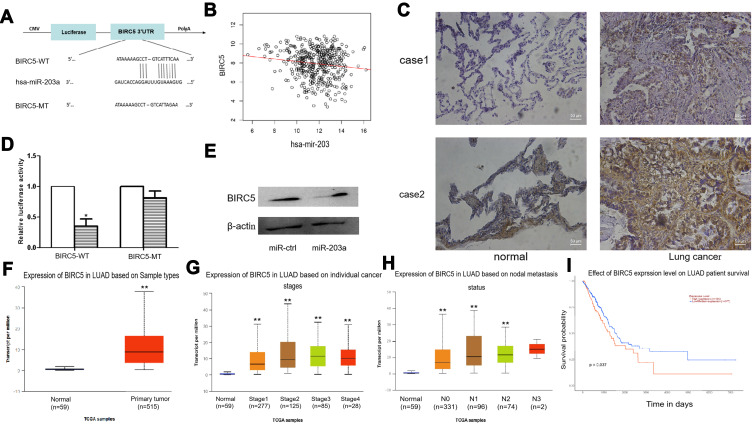
We found that BIRC5 had a potential miR-203a binding site in its 3ʹ-untranslated region (UTR) using miRNA target prediction programs (Figure 5A). Correlation analysis of miR-203 and BIRC5 was performed by LinkedOmics14 and their expression levels were found to be negatively correlated (Figure 5B). We examined protein abundance of BIRC5 by immunohistochemical (IHC) staining, and the results showed that BIRC5 abundance was higher in lung cancer tissues than in normal tissues (Figure 5C).
Figure 5.
BIRC5 is a direct target of miR-203a. (A) miR-203a has the binding sites of the 3′-UTR of BIRC5. (B) The expression correlation analysis of miR-203 and BIRC5 was analyzed by Linkedomics. (C) IHC assay was performed to detect the protein expression of BIRC5 in lung cancer tissues. (D) The luciferase assay was performed with pre-miR-203a co-transfected with pGLO-BRIC5 wild-type or pGLO-BIRC5 mutant vector in HEK293 cells, pre-miR-203a co-transfected with pGLO was used as control. (E) The protein expression levels of BIRC5 were detected by Western blot after transfection with pre-miR-203a or miR-control. (F) UALCAN was used to analyze the expression of BIRC5 in Lung adenocarcinoma. (G) UALCAN was used to analyze the expression of BIRC5 in different lung adenocarcinoma stages. (H) Expression of BIRC5 in lung adenocarcinoma based on nodal metastasis status analyze by UALCAN. (I) Survival of lung adenocarcinoma patients with difference of BIRC5 expression (*P<0.05, **P<0.01).
We then performed the dual-luciferase reporter assays to confirm whether BIRC5 is a direct target of miR-203a. Luciferase activity in the pre-miR-203a and wild-type (wt) BIRC5 3ʹ-UTR co-transfection group was significantly reduced compared with that in the control group, the control group is pre-miR-203a and pmirGLO vector co-transfection. Meanwhile, luciferase activity was not altered in the miR-203a and mutant BIRC5 3ʹ-UTR co-transfection group compared with that in control group (Figure 5D). Additionally, Western blotting was conducted to measure protein expression for BIRC5 after A549 cells were transfected with pre-miR-203a or miR-ctrl. Pre-miR-203a transfection in A549 cells downregulated BIRC5 expression. Furthermore, Western blotting demonstrated that miR-203a overexpression reduced BIRC5 protein abundance in A549 cells (Figure 5E). Together, these results suggested that miR-203a may act as a tumour suppressor and prevent lung cancer progression.
Next, we used UALCAN15 to analyse the expression of BIRC5 in lung adenocarcinomas, and found that expression of BIRC5 was higher in tumours than in normal tissue. Additionally, expression of BIRC5 was higher in stage I–IV LUAD than controls, expression increased with increasing lymph node invasion, and high expression of BIRC5 correlates with worse prognosis (Figure 5F–I).
Silencing of BIRC5 Suppresses Proliferation and Induces Apoptosis in Lung Cancer Cells
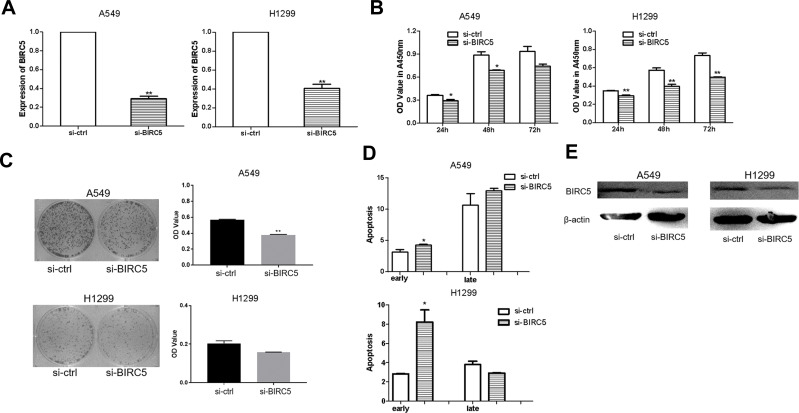
The effectiveness of si-BIRC5 was examined by qRT-PCR in A549 and H1299 cells. We observed that expression of BIRC5 was decreased after transfection with si-BIRC5 (Figure 6A). To determine whether BIRC5 knockdown contributed to growth inhibition, we performed CCK-8 assays and the colony formation assays in A549 and H1299 cells after transfection with si-BIRC5. BIRC5 knockdown had a significant inhibitory effect in both A549 and H1299 cells (Figure 6B and C), as evidenced by the cell apoptosis assays which showed that si-BIRC5 increased early apoptosis in A549 and H1299 cells (Figure 6D). Western blotting demonstrated that BIRC5 protein expression was reduced in A549 and H1299 cells transfected with si-BIRC5 (Figure 6E). These findings demonstrated that silencing of BIRC5 inhibited cell growth and induced apoptosis in lung cancer cells.
Figure 6.
Silencing of BIRC5 suppresses proliferation and induces apoptosis in lung cancer cells. (A) the expression of BIRC5 was detected after transfected with si-BIRC5 comparing with si-ctrl in A549/H1299 cells. (B) CCK-8 assay was to detect the effect of silencing of BIRC5 on A549/H1299 cells transfected with si-BIRC5 or si-ctrl. (C) Colony formation was performed to measure the effect of silencing of BIRC5 on A549/H1299 cells transfected with si-BIRC5 or si-ctrl. (D) Apoptosis was detected in A549/H1299 cells after transfection with si-BIRC5 or si-ctrl. (E) Western blot was used to detect the BIRC5 protein expression levels in A549/H1299 cells which transfection with si-BIRC5or si-trl (*P<0.05, **P<0.01).
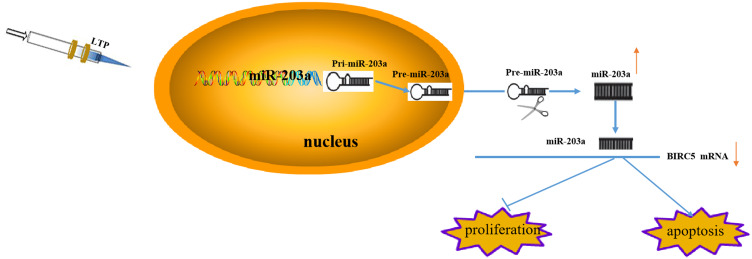
LTP Mediates miR-203a Suppression of Proliferation and Induces Apoptosis of Lung Cancer Cells by Targeting BIRC5
LTP suppressed proliferation and promoted apoptosis in lung cancer cells, and induced expression of miR-203a. Activated miR-203a suppressed growth and induced apoptosis in lung cancer cells by targeting BIRC5 (Figure 7).
Figure 7.
LTP mediates miR-203a/BIRC5 axis suppress proliferation and induces apoptosis of lung cancer cells.
Discussion
Medical applications for LTP are increasing since studies have shown that it can be used as an additional cancer therapy. Our present study employed an APPJ with multiple electrodes to produce plasma, and LTP had obvious effects on A549 and H1299 lung cancer cells. Specifically, LTP promoted apoptosis and suppressed proliferation of lung cancer cells. We further investigated the mechanism of LTP in lung cancer progression and found that miR-203a was increased when A549 or H1299 cells were treated with LTP, suggesting that miR-203a may play a key role in growth inhibition of lung cancer cells by LTP.
Numerous studies have shown that miRNAs play important roles during lung cancer progression. However, the exact role of miRNA dysregulation in the pathogenesis of lung cancer requires further investigation. Our results showed that miR-203a overexpression suppressed proliferation and promoted apoptosis in A549 and H1299 cells. The miR-203a may be a potential tumour suppressor gene in lung cancer. The miRNAs interact with the 3ʹ-UTR of the target gene mRNA to regulate mRNA degradation or inhibition. Through bioinformatics analysis and dual-luciferase reporter assays, we demonstrated that miR-203a could target BIRC5, a member of the inhibitor of apoptosis (IAP) gene family, which consists of BIRC1–8. The BIRC5 gene, also known as survivin, is associated with most common human cancers, including human endometrial cancer,16 bladder cancer,17,18 hepatocellular carcinoma,19 breast cancer20 and ovarian cancer.21 BIRC5 may play a critical role in regulating the cell cycle, proliferation, progression and angiogenesis in cancer cells.22 Thus, BIRC5 may be a potential target for cancer treatment.23,24 Herein, we found that si-BIRC5 inhibited proliferation in both A549 and H1299 cells.
Our study demonstrated inhibition of proliferation by LTP in lung cancer cells. Furthermore, we found that miR-203a was increased after lung cancer cells were treated with LTP. We focused on the miR-203a/BIRC5 axis in LTP-treated cells and revealed that miR-203a targets BIRC5 to induce apoptosis in lung cancer cells. Thus, APPJ may be an attractive anti-cancer treatment method. Our findings demonstrated that the miR-203a/BIRC5 axis was affected by LTP, which may explain why LTP suppressed lung cancer cell growth.
Acknowledgments
This work was supported by Postdoctoral Science Foundation of China (no. 2017M623195).
Disclosure
The authors declare there is no conflict of interest in this work.
References
- 1.Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.1016/S0025-6196(11)60735-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiraki T, Gobara H, Iguchi T, et al. Radiofrequency ablation for early-stage nonsmall cell lung cancer. Biomed Res Int. 2014;2014:152087. doi: 10.1155/2014/152087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd G, Friedman G, Jafri S, et al. Gas plasma: medical uses and developments in wound care. Plasma Process Polym. 2010;7:194–211. doi: 10.1002/ppap.200900097 [DOI] [Google Scholar]
- 4.McCombs GB, Darby ML. New discoveries and directions for medical, dental and dental hygiene research: low temperature atmospheric pressure plasma. Int J Dent Hyg. 2010;8(1):10–15. doi: 10.1111/j.1601-5037.2009.00386.x [DOI] [PubMed] [Google Scholar]
- 5.Miyamoto K, Ikehara S, Takei H, et al. Red blood cell coagulation induced by low-temperature plasma treatment. Arch Biochem Biophys. 2016;605:95–101. doi: 10.1016/j.abb.2016.03.023 [DOI] [PubMed] [Google Scholar]
- 6.Rothschild SI. microRNA therapies in cancer. Mol Cell Ther. 2014;2:7. doi: 10.1186/2052-8426-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Xu L, Zheng J, et al. MiR-101 inhibits ovarian carcinogenesis by repressing the expression of brain-derived neurotrophic factor. FEBS Open Bio. 2017;7(9):1258–1266. doi: 10.1002/2211-5463.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye G, Huang K, Yu J, et al. MicroRNA-647 targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer. Theranostics. 2017;7(13):3338–3353. doi: 10.7150/thno.20512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Tong K, Yu J. MicroRNA-130a is upregulated in colorectal cancer and promotes cell growth and motility by directly targeting forkhead box F2. Mol Med Rep. 2017;16(4):5241–5248. doi: 10.3892/mmr.2017.7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Ding L, Hu Q, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16(1):141. doi: 10.1186/s12943-017-0710-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Liao Z, Cai M, Zhang G. MicroRNA-320a downregulation mediates human liver cancer cell proliferation through the Wnt/beta-catenin signaling pathway. Oncol Lett. 2017;13(2):573–578. doi: 10.3892/ol.2016.5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zhao Z, Yang Y, et al. MiR-99b-5p and miR-203a-3p function as tumor suppressors by targeting IGF-1R in gastric cancer. Sci Rep. 2018;8:10119. doi: 10.1038/s41598-018-27583-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Tong D, Guo Q, et al. HOXD3 targeted by miR-203a suppresses cell metastasis and angiogenesis through VEGFR in human hepatocellular carcinoma cells. Sci Rep. 2018;8:2431. doi: 10.1038/s41598-018-20859-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res.2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai Z, Yin L, Zhou X, et al. Inhibition of survivin reduces cell proliferation and induces apoptosis in human endometrial cancer. Cancer. 2006;107:746–756. doi: 10.1002/cncr.22044 [DOI] [PubMed] [Google Scholar]
- 17.Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. doi: 10.1186/s12943-016-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui X, Shen D, Kong C, et al. NF-kappaB suppresses apoptosis and promotes bladder cancer cell proliferation by upregulating survivin expression in vitro and in vivo. Sci Rep. 2017;7:40723. doi: 10.1038/srep40723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao L, Li C, Shen S, et al. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer. 2013;13:82. doi: 10.1186/1471-2407-13-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han W, Cao F, Gao XJ, et al. ZIC1 acts a tumor suppressor in breast cancer by targeting survivin. Int J Oncol. 2018;53(3):937–948. doi: 10.3892/ijo.2018.4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G, Wang Q, Gu Q, et al. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8(55):94666–94680. doi: 10.18632/oncotarget.21863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin: molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 2007;67(13):5999–6002. doi: 10.1158/0008-5472.CAN-07-0494 [DOI] [PubMed] [Google Scholar]
- 23.Lyu H, Huang J, He Z, Liu B. Epigenetic mechanism of survivin dysregulation in human cancer. Sci China Life Sci. 2018;61(7):808–814. doi: 10.1007/s11427-017-9230-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70. doi: 10.1038/nrc2293 [DOI] [PubMed] [Google Scholar]