Fig. 2.
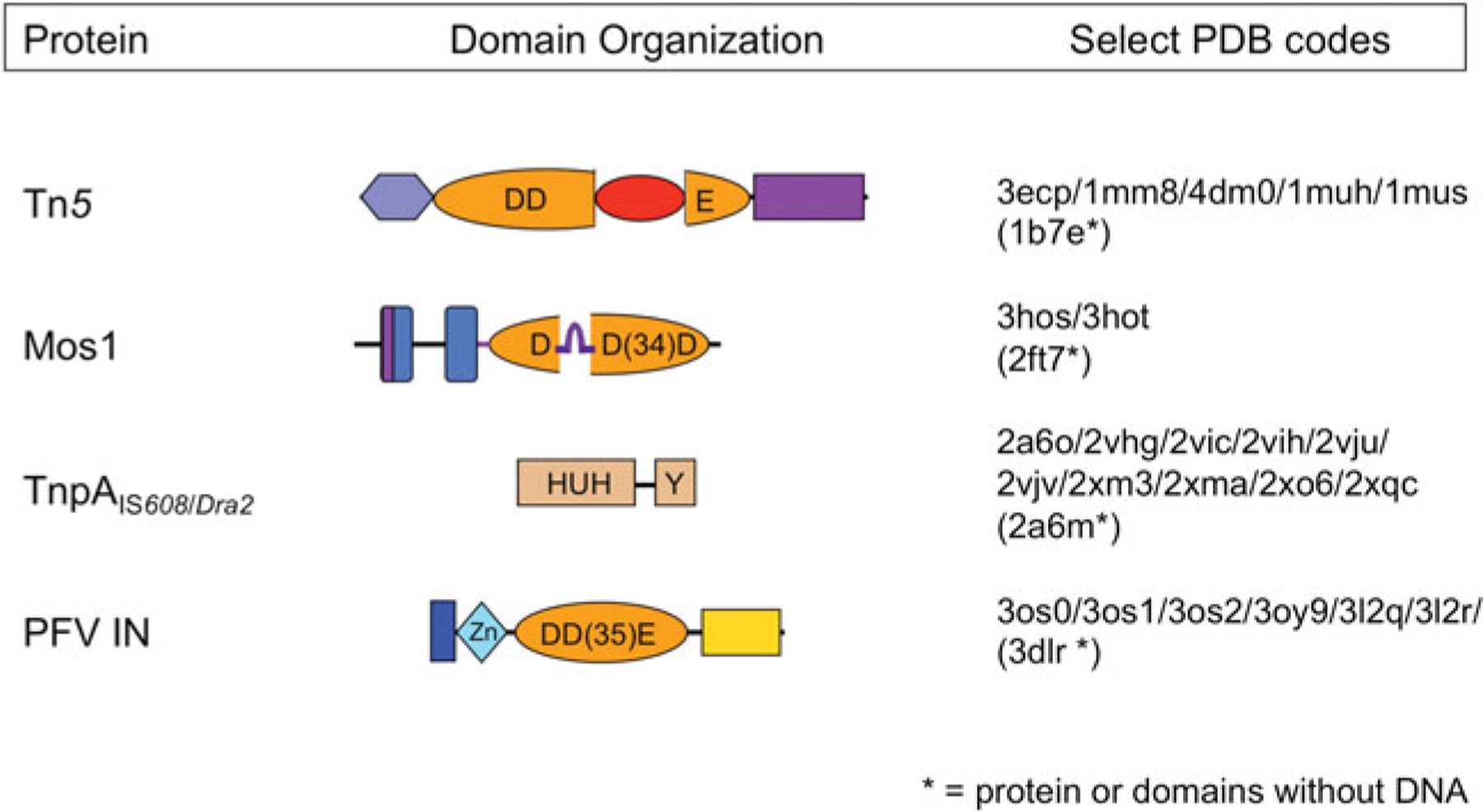
Domain organization of select transposases. The four transposases for which there are high resolution transpososome structures are shown schematically, with DNA-binding domains in shades of blue and nuclease catalytic domains in shades of orange. Regions important for multimerization are shown in purple. The Tn5 and Mos1 transposases, and PFV integrase (PFV IN) have RNaseH-like catalytic domains which use three acidic residues (DDD or DDE) to coordinate two divalent metal ions required for catalysis. The TnpA ssDNA transposases use an HUH nuclease domain to coordinate a single metal ion which, in conjunction with a catalytic tyrosine, comprises the enzyme active site. The four beta-strand insertion into the Tn5 catalytic domain is shown in red, and the ‘clamp-loop’ of Mos1, inserted relative to the standard RNaseH topology between beta strands 1 and 2, is shown in purple.
