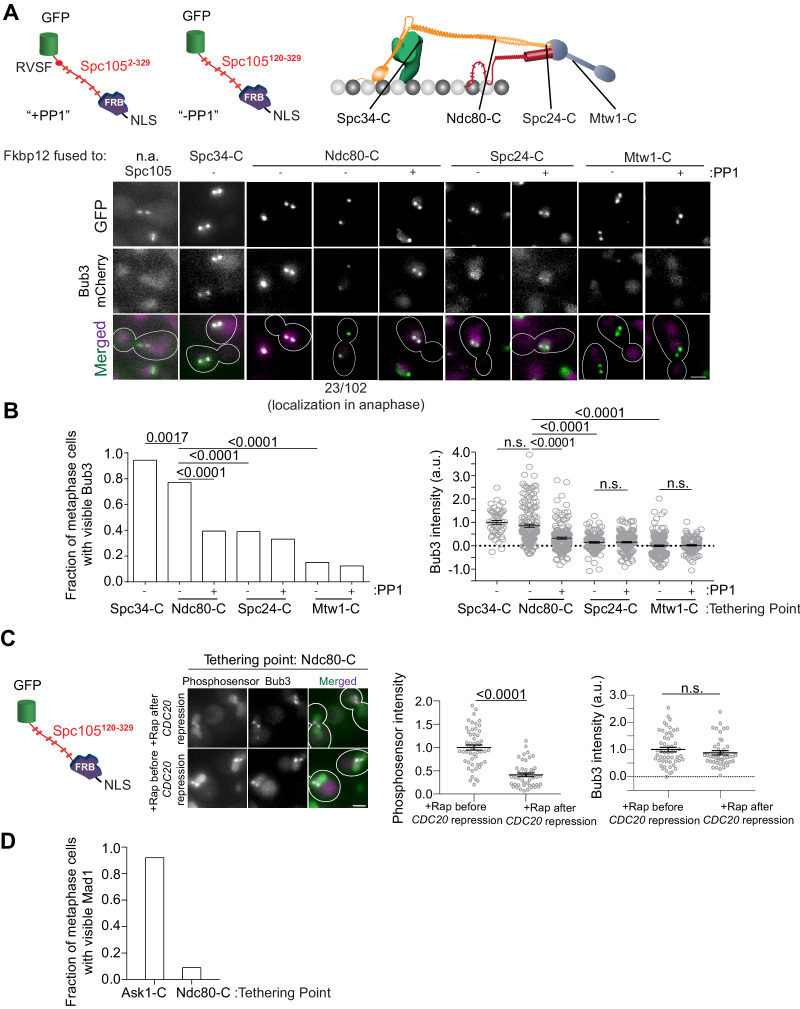
Figure 3. A gradient of Mps1 kinase activity continues to phosphorylate MELT motifs after chromosome biorientation.
(A) Top left: Cartoons show schematics of the two Spc105 phosphodomains tethered to kinetochore subunits using the rapamycin-induced dimerization of Fkbp12 and Frb domains. Top right: The kinetochore subunits (Spc34-C, Ndc80-C, Spc24-C and Mtw1-C) at which, the phosphodomains were tethered by rapamycin induced dimerization. Bottom: Micrographs show representative images of cycling cells wherein the phosphodomain is tethered to the C-terminus of the indicated kinetochore subunit. (n.a. - Not Applicable). (B) Left: The bar plot shows the scoring of cells with bioriented kinetochore clusters based on whether or not they visibly recruit Bub3-mCherry. (n = 71 for Spc34-C, 109 and 94 for Ndc80-C, 95 and 103 for Spc24-C, 106 and 122 for Mtw1-C, accumulated from two experimental and biological repeats (whenever possible). Right: The scatter plot on the right shows the quantification of Bub3-mCherry fluorescence from all cells with bioriented kinetochore clusters. (mean+ s.e.m. n = 47 for Spc34-C, 159 and 153 for Ndc80-C, 114 and 154 for Spc24-C, 173 and 104 for Mtw1-C, accumulated from two experimental and biological repeats wherever possible). n.s. Not significant. (C) Micrographs show the recruitment of Bub3-mCherry by the phosphosensor tethered at the Ndc80 C-terminus in cycling (before CDC20 repression) and metaphase arrested cells (after CDC20 repression). Scatterplots show the relative fluorescence signal from the GFP-tagged phosphosensor and Bub3-mCherry respectively. (mean+ s.e.m. n = 52 and 45 for rapamycin treated samples before and after Cdc20 depletion from two technical replicates). n.s. Not significant. (D) Bar graph shows fraction of cells visibly recruiting Mad1-mCherry as shown in the micrographs (n = 242 and 142 for Ask1-C and Ndc80-C respectively).