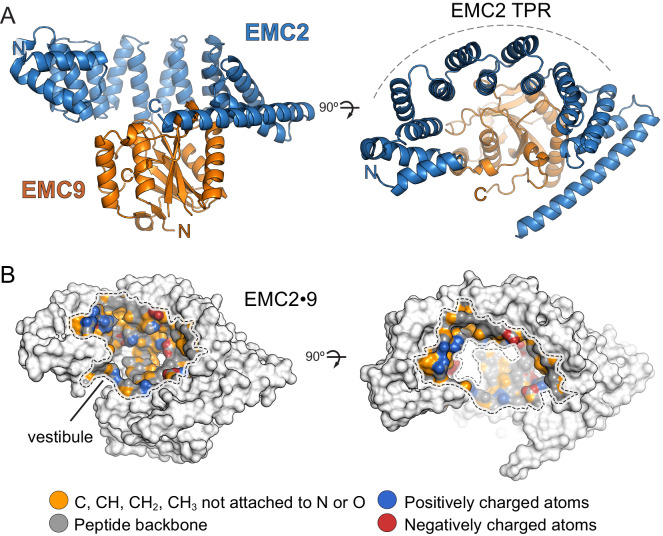
Figure 2. Structure of the EMC2•EMC9 complex.
(A) Crystal structure of the EMC2•EMC9 heterodimer (PDB: 6Y4L). The heterodimer consists of EMC2 (residues 11–274), depicted in blue, and EMC9 (residues 1–200), depicted in orange. The TPR-repeat motif of EMC2 is indicated. (B) Physicochemical properties of a vestibule in the EMC2•EMC9 complex. Surface rendering of crystal structures coloured according to chemical properties (Hagemans et al., 2015).
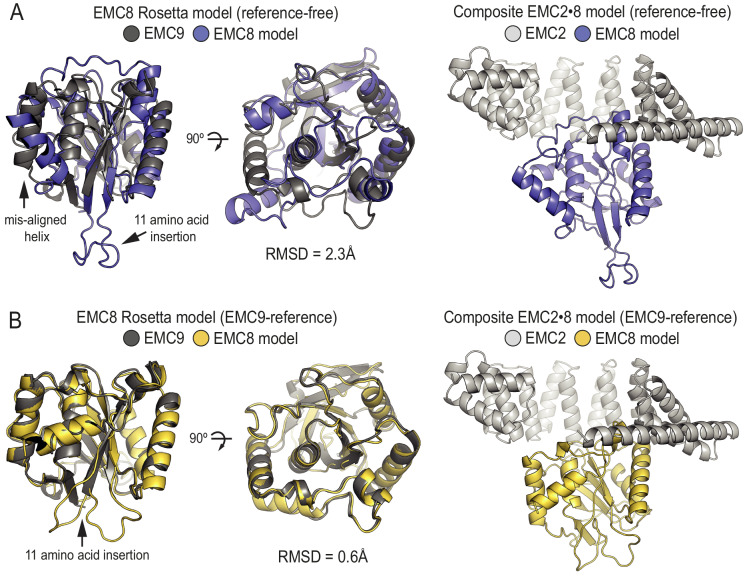
Figure 2—figure supplement 1. Protein structure prediction confirms structural homology between EMC9 and EMC8.
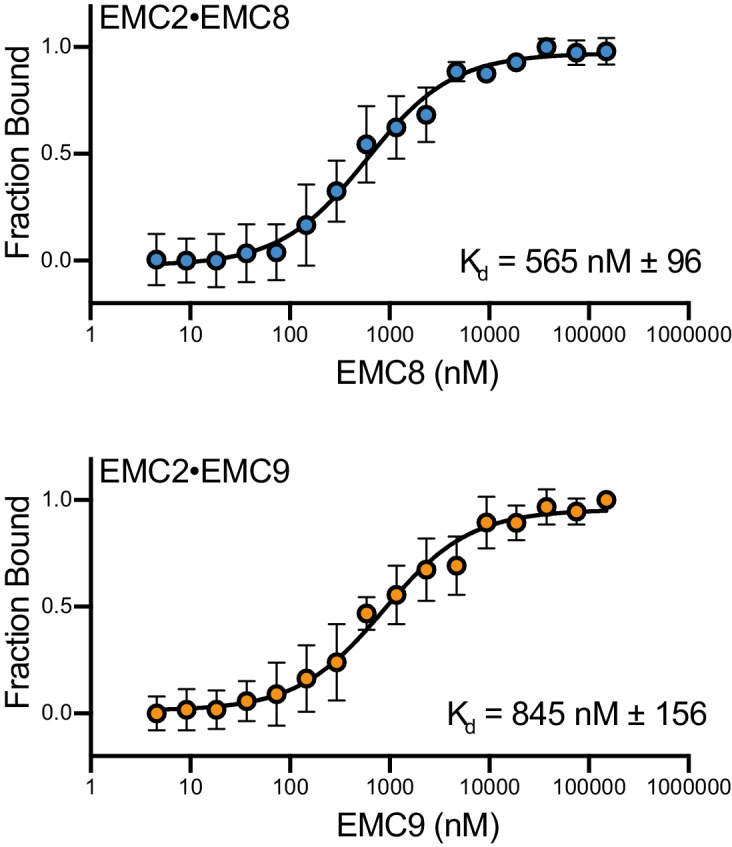
Figure 2—figure supplement 2. EMC8 and EMC9 have similar affinities for EMC2.