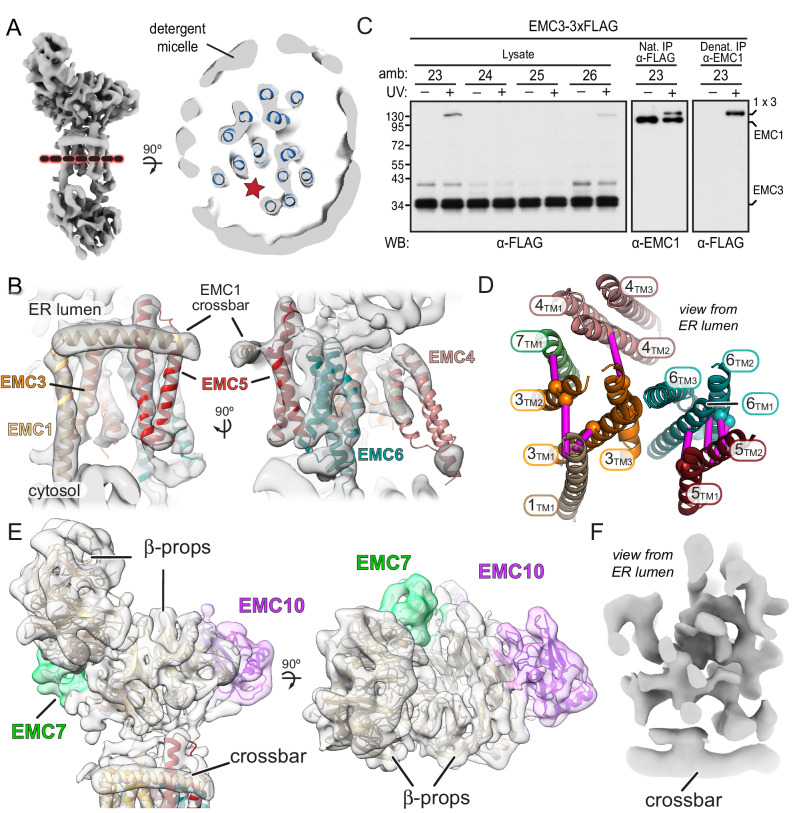
Figure 5. A composite model of EMC’s membrane and lumenal domains.
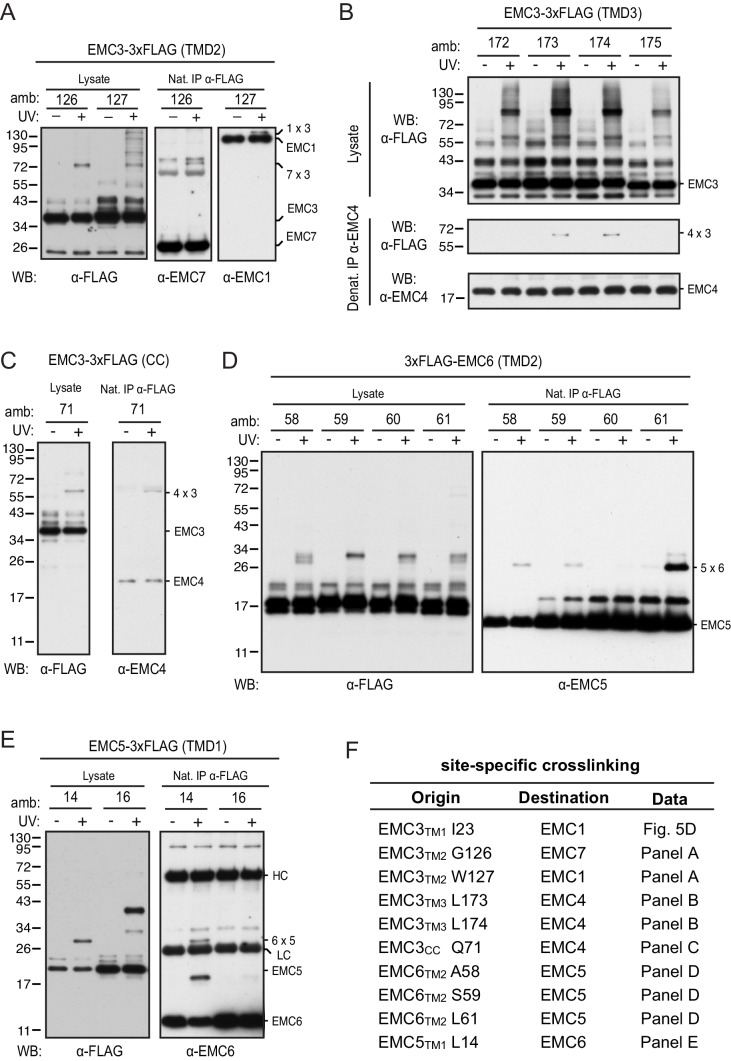
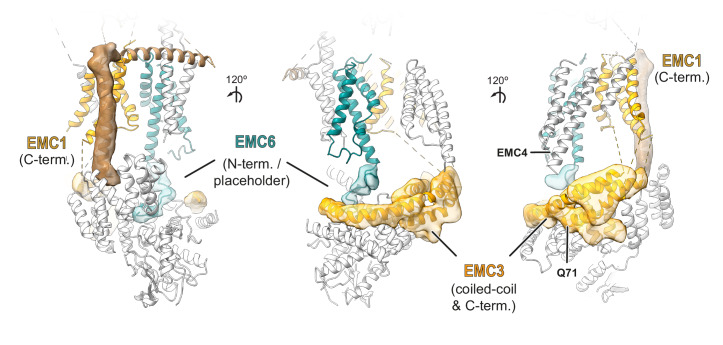
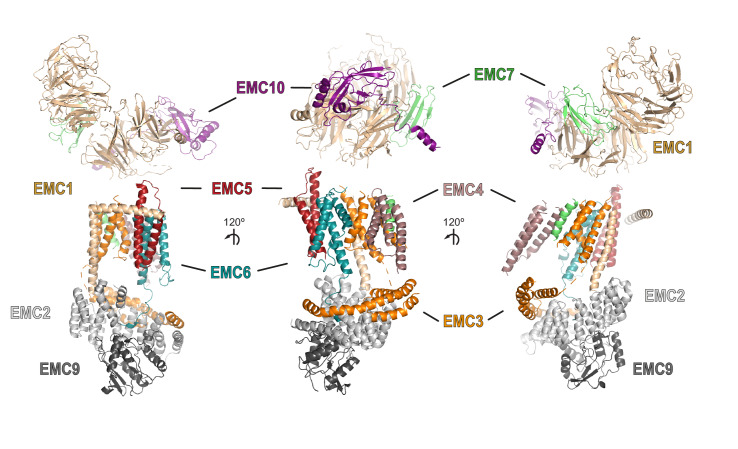
(A) Cross section through the TMD region of the cryo-EM map of EMC (light grey) identifies 13 helix-like densities (dark blue) that define an intramembrane groove (red star) open to the lipid bilayer. Density corresponding to the annular detergent micelle is indicated. (B) Ab initio trRosetta models fitted into cryo-EM density for the EMC membrane domain are shown for EMC1 (wheat), EMC3 (orange), EMC4 (light pink), EMC5 (red), EMC6 (teal), and EMC7 (green). The horizontal density at the membrane-lumen interface termed the ‘crossbar’ is assigned to an amphipathic helix in EMC1. (C) EMC3-3xFLAG constructs containing amber codons (amb.) at the indicated positions were expressed together with an amber suppressor tRNA and cognate aminoacyl-tRNA synthetase that accepts the UV-activated crosslinking amino acid 3'-azibutyl-N-carbamoyl-lysine (AbK). Cells were left untreated or irradiated with UV and analyzed by immunoblotting for EMC3-3xFLAG. A prominent UV-dependent crosslink is seen from position 23 and to a lesser extent, position 26. Native FLAG immunoprecipitation (IP) recovers EMC1 (indicating that EMC3-3xFLAG is incorporated into EMC), which shifts with UV. Denaturing EMC1 IP confirms the crosslinked product contains both EMC1 and EMC3. (D) TMD helices positioned based on docking of ab initio models overlayed with AbK-mediated crosslinks (see Figure 5—figure supplement 3). The positions where AbK was incorporated are shown as spheres, with magenta lines showing the closest point of the target protein in the model. (E) Composite model of the EMC lumenal domain generated by ab initio modelling in trRosetta and real-space refinement in PHENIX. Cryo-EM density has been colored according to subunit identity and the composite lumenal domain model accounts for almost all the lumenal EM density. (F) Cross-section through EMC at the plane of the membrane-lumen interface illustrating that a pore is not evident across the membrane. All EM data visualized in UCSF ChimeraX with EM maps contoured at 0.15 (panel A) and 0.21 (panels B, E, and F) with hide dust setting of 10. Panel D was generated in PyMOL.
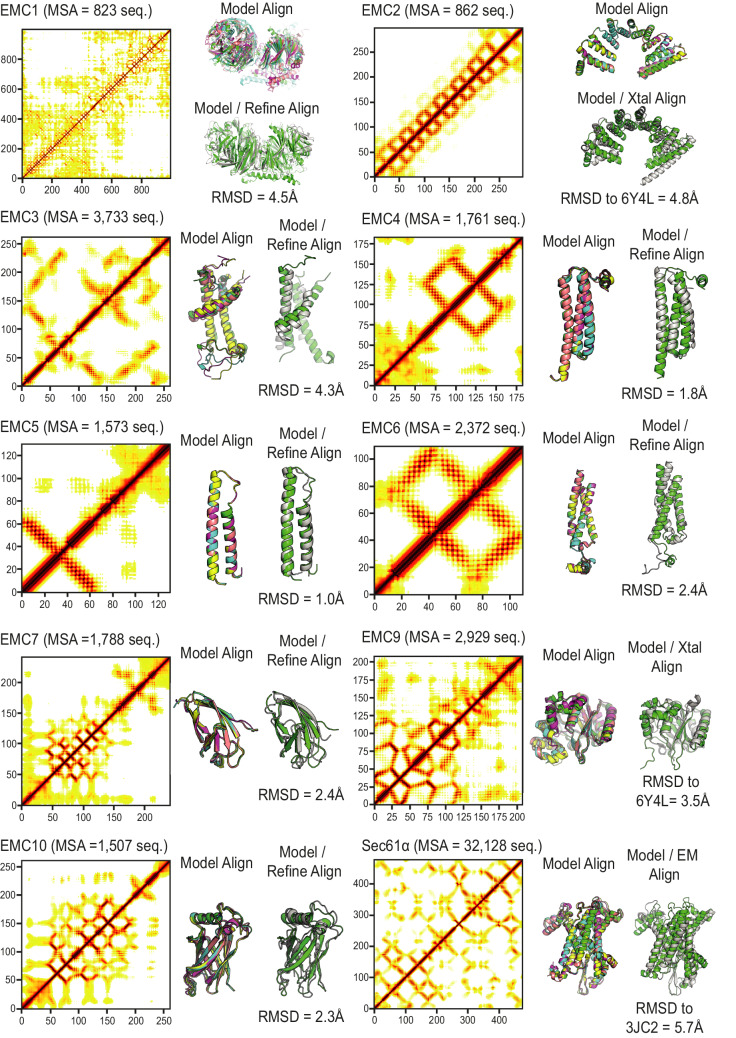
Figure 5—figure supplement 1. Ab initio prediction of EMC subunit structure and flexible fitting using real-space refinement.
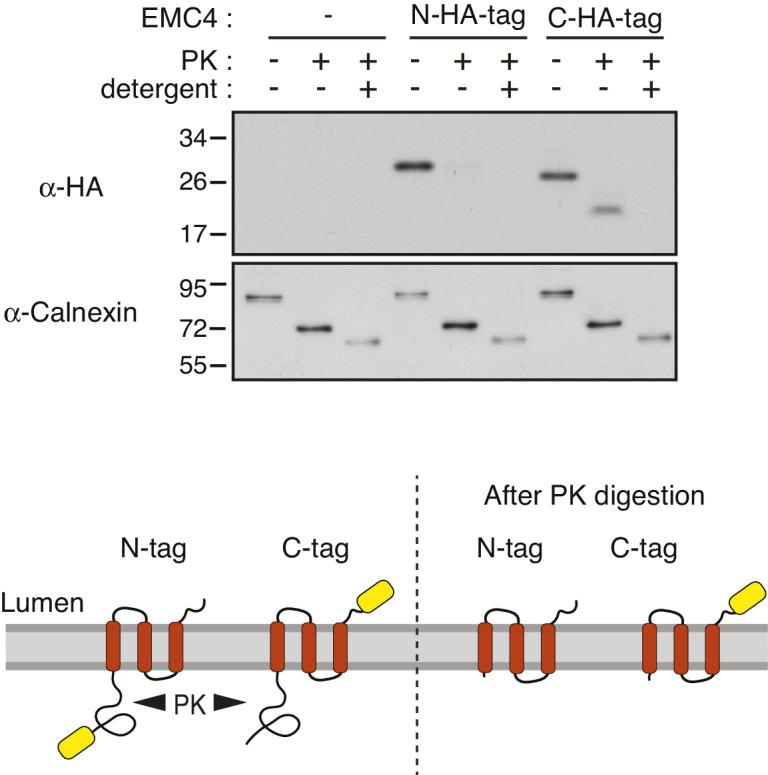
Figure 5—figure supplement 2. Protease-protection analysis of EMC4 topology.