Abstract
The FDA lists 22 medications as clinical inhibitors of CYP2D6, with classifications of strong, moderate, and weak. It is accepted that strong inhibitors result in nearly null enzymatic activity, but reduction caused by moderate and weak inhibitors is less well characterized. The objective was to identify if the classification of currently listed FDA moderate and weak inhibitors is supported by publicly available primary literature. We conducted a literature search and reviewed product labels (PLs) for AUC-fold changes caused by inhibitors in humans and identified 89 inhibitor-substrate pairs. Observed AUC-fold change of the substrate was used to create an observed inhibitor classification per FDA-defined AUC-fold change thresholds. We then compared the observed inhibitor classification with the classification listed in the FDA Table of Inhibitors. We found 62% of the inhibitors within the pairs matched the listed FDA classification. We explored reasons for discordance and suggest modifications to the FDA table of clinical inhibitors for cimetidine, desvenlafaxine, and fluvoxamine.
Keywords: CYP, Drug-Drug Interactions, Inhibitors, Precision medicine
INTRODUCTION
Cytochrome P450 family 2 subfamily D member 6 (CYP2D6) contributes to 2% and 0.7% of the overall hepatic and intestinal cytochrome P450 enzyme, respectively,(1) and is estimated to be responsible for metabolism of 25% of commonly prescribed drugs (e.g., antidepressants, antipsychotics, opioids, beta blockers).(2) Significant interpatient variability in pharmacokinetics, pharmacodynamics, and clinical outcomes have been attributed to interpatient variability of CYP2D6 function.(3) An important source of variability in CYP2D6 function arises from genetic variability, including variability that leads to no function, decreased function and increased function.(4) The genetic variation is commonly categorized into phenotypes, which include poor metabolism (PM), intermediate metabolism (IM), normal metabolism (NM), and ultra-rapid metabolism (UM).
CYP2D6 not only metabolizes a large number of drugs but is also susceptible to clinically-relevant inhibition by a number of commonly used drugs.(5) This can lead to a clinical phenotype that does not align with the genetically-defined phenotype, a process often called phenoconversion. For example, the strong CYP2D6 inhibitor paroxetine can phenoconvert an individual who is genetically a NM to a PM.(6) CYP2D6 inhibitors use is prevalent and has been observed to cause phenoconversion in 17% and 25% of individuals from two different studies.(7, 8) This is of great clinical importance as serious adverse drug reactions can occur from CYP2D6-mediated drug-drug interactions (DDIs). In instances where CYP2D6 metabolizes a drug to its inactive form, such as with atomoxetine to 4-hydroxyatomoxetine, phenoconversion may lead to higher rates of adverse effects. In contrast, when CYP2D6 metabolizes a drug to its active form, such as with codeine to morphine, phenoconversion can reduce drug effectiveness. It has been suggested that consideration of both CYP2D6 genotype along with CYP2D6 drug interactions to infer the CYP2D6 clinical phenotype is a preferred approach for precision medicine.(9, 10)
Potential mechanisms to understanding inter-individual CYP2D6 activity are estimation of CYP2D6 function by phenotyping via a probe drug (e.g., dextromethorphan) and genotyping CYP2D6. However, there are limitations with each. Phenotyping is not routinely performed clinically, and genotype data may be insufficient to predict the clinical phenotype if CYP2D6 inhibitors are being taken concomitantly. Therefore, clinicians tend to rely on the Food and Drug Administration (FDA) labeling and drug-drug interaction resources as guidance for defining clinically relevant enzyme inhibition drug-drug interactions. As primary literature and many practicing clinicians would indicate, current drug-drug interaction tools are inconsistent with one another regarding drug-drug interaction classification or recommendations. Patel et al. found only 35% to 70% consistency between commonly used drug-drug interaction resources.(11)
While there are multiple resources for classifying drugs as inhibitors, the FDA Table of Inhibitors is commonly considered to be the authoritative resource. (12) The FDA lists at least 22 medications as strong, moderate, or weak inhibitors. Medications classified as strong are largely consistent among resources.(12, 13) It is well accepted that strong inhibitors decrease CYP enzymatic activity to nearly null, thus essentially converting an individual with a NM or IM genotype to a PM.(14, 15) However there may be less confidence in moderate and weak inhibitors because of uncertainty about or variability in the magnitude of CYP2D6 inhibition they cause based on published literature. If clinicians seek to consider drug-drug-gene interactions (DDGIs) in their clinical decision-making, they should be confident with applying phenoconversion in practice, as it could change drug therapy choices.
The objective of this project was to identify whether the classifications of inhibitors listed as moderate and weak in the FDA Table of Inhibitors is supported by publicly available primary literature by examining data supporting the effects of CYP2D6 inhibitors on the area under the plasma concentration-time curve (AUC) on CYP2D6 substrates in humans.
METHODS
A scoping review was conducted using FDA product labels (PLs) and PubMed (inception to December 12th, 2018). Search terms were: (moderate OR weak inhibitors) AND (sensitive OR moderate-sensitive substrates) AND (pharmacokinetics OR CYP2D6 OR drug interaction) with individual drug names inserted per listed classification in the FDA Tables of Inhibitors and Substrates. Table S1 displays the drugs listed in the FDA Tables of Inhibitors and Substrates at the time of this search. FDA PLs were obtained online from the FDA website.(https://www.accessdata.fda.gov/scripts/cder/daf/).
Resulting articles were uploaded to Rayyan,(16) an application used to manage the article screening process. Four authors were involved in the screening process (E.J.C., D.M.S., B.Q.D., L.G.K.) and rotated roles. The abstract for each article was independently screened for inclusion by two authors. Disagreements were resolved by a third author. Abstracts that passed screening were simultaneously assessed for eligibility and data were collected if included. Inclusion criteria for articles were: conducted in humans and reported AUC of the substrate with and without the inhibitor. Case reports were excluded. FDA PLs were also assessed for eligibility and excluded if the inclusion criteria were not met. FDA PLs and articles are hereafter referred to as records.
Data collected from each record included study design, intervention, study population, CYP2D6 phenotype (if determined), CYP2D6 phenotype assessment method (if performed), inhibitor, substrate, dose, number of doses administered, AUC of substrate alone, and AUC of substrate with inhibitor. Additionally, any information regarding inhibition was noted when PLs were reviewed. Fold-change in substrate AUC was calculated for each inhibitor-substrate pair (i.e., AUC of substrate with inhibitor divided by AUC of substrate alone). Some records reported AUC values at multiple doses, time points, or reported values by CYP2D6 phenotype. Therefore, one record may have resulted in multiple inhibitor-substrate pairs. All data collection was performed and verified by two authors and any disagreements were adjudicated by a third.
Per the FDA, CYP2D6 strong, moderate, and weak inhibitors are drugs that increase the AUC of CYP2D6 sensitive index substrates by ≥5-fold, ≥2 to <5-fold, and ≥1.25 to <2-fold, respectively.(12) We determined observed CYP2D6 inhibitor classification utilizing the same thresholds, but regardless of classification of substrate (i.e., moderate sensitive or sensitive). AUC fold-change <1.25 was classified as insignificant. As a measure of sensitivity, we also assessed the inhibitor classifications by limiting to sensitive substrates. The primary outcome was selected to examine how often the observed inhibitor classification matched the inhibitor’s classification listed on the FDA Table of Inhibitors, herein referred to as concordance. The secondary outcome was selected to evaluate study criteria to identify factors associated with concordance and discordance of the observed inhibitor classification with the classification listed in the FDA Table of Inhibitors. Study criteria evaluated substrates, if the inhibitor inhibits multiple metabolic pathways, total daily dose of inhibitor, if the inhibitor was given as a single dose or at steady state, and phenotype of participants. Inhibitors included in the secondary outcome were those with more than one inhibitor-substrate pair present. These study criteria informed the author interpretations. Substrates metabolized by multiple CYP enzymes and given with a drug that inhibited those multiple pathways may exhibit inflated increases in AUC. Drugs were defined to affect multiple pathways if a source listed the drug as an inhibitor or substrate for a second pathway.(12, 13, 17) These CYP2D6 inhibitors were listed as inhibitors of other pathways, with the other pathways shown in parentheses: fluvoxamine (CYP2C19), cimetidine (CYP1A2), sertraline (CYP2C9), and ritonavir (CYP3A). These CYP2D6 substrates were listed as substrates of other pathways: amitriptyline (CYP1A2, CYP2C9, CYP2C19), dextromethorphan (CYP3A), imipramine (CYP1A2, CYP2C19), and propranolol (CYP1A2).
RESULTS
Literature Search
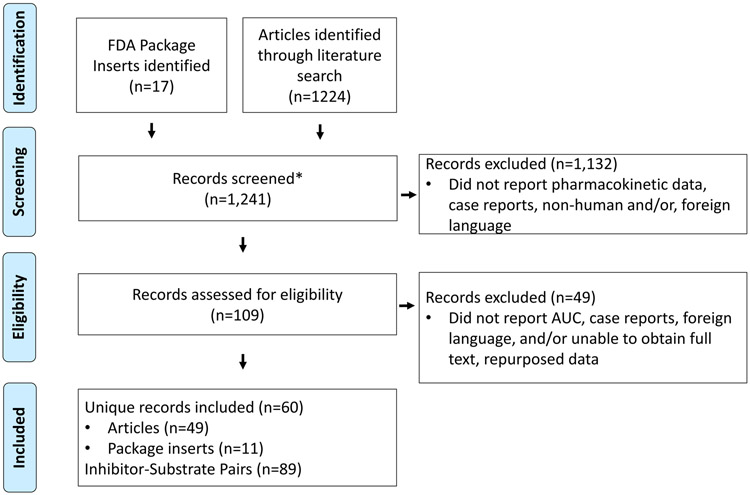
The PubMed search yielded 1,224 articles, which were uploaded into Rayyan.(16) Author screening had a 94% agreement rate. PLs of the 17 drugs classified as a moderate or weak in the FDA Table of Inhibitors were assessed for eligibility. Details are shown in Figure 1, in total 60 records were included, resulting in 89 inhibitor-substrate pairs. References for all records can be found in Table S2.
Figure 1: Flow Diagram.
Product labels for the 17 inhibitors classified as moderate or weak per the FDA Table of Inhibitors were collected and articles were identified through a literature search. They were both then screened (e.g., abstract review) for eligibility. Records lacking exclusion criteria were assessed in more detail (e.g., full article review) for eligibility. Records were included if they were conducted in humans and reported AUC of substrate with and without the inhibitor. Each inhibitor-substrate pair that had a report of AUC was counted as one pair.
*No duplicates records present
Concordance with Classifications Per the FDA Table of Inhibitors
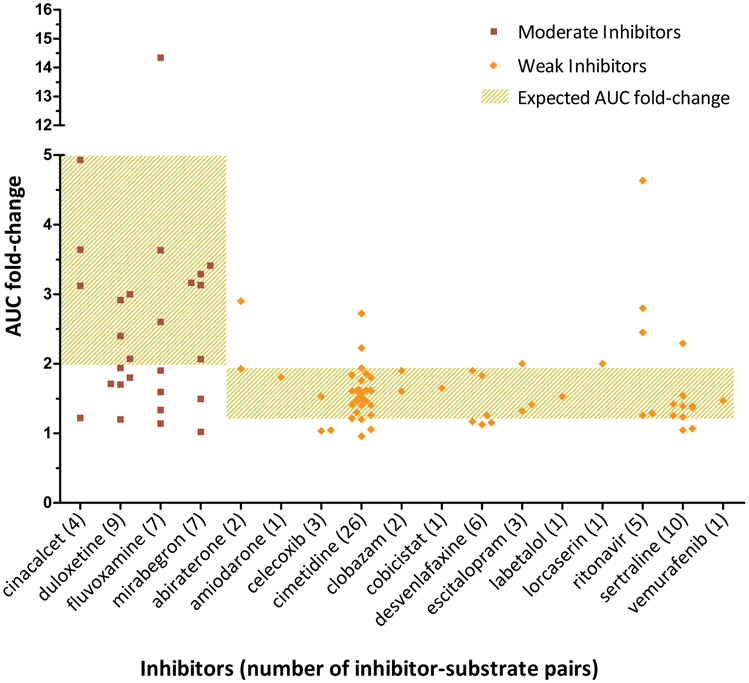
The overall concordance of observed inhibitor classifications with the classification per the FDA Table of Inhibitors was 62% (55 of 89 inhibitor-substrate pairs). Concordance by classification of inhibitor (moderate or weak) with classification of substrate (sensitive or moderate sensitive) can be seen in Table 1. The pairs involving a weak inhibitor had the highest concordance, 75% with a moderate sensitive substrate and 57% with a sensitive substrate. A visual representation of concordance can be seen in Figure 2, where the AUC fold-change of the substrate is plotted by inhibitor. Pairs that did not fall within the expected range, typically did not meet the defined threshold for the inhibitor classification. For example, for the following inhibitors, > 50% of their respective pairs did not meet the threshold for their respective classification; duloxetine (moderate), fluvoxamine (moderate), celecoxib (weak), and desvenlafaxine (weak). When concordance assessment was limited to only sensitive substrates, the concordance was 56% (27 of 48 inhibitor-substrate pairs).
Table 1:
Concordance by Classification per FDA Table of Inhibitors
| Inhibitorsa | Substrates | Concordance, n (%) (n= 89 pairs) |
|---|---|---|
| All | All | 55 of 89 (62) |
| Moderate | Sensitive | 10 of 18 (56) |
| Moderate | Moderate sensitive | 4 of 9 (44) |
| Weak | Sensitive | 17 of 30 (57) |
| Weak | Moderate sensitive | 24 of 32 (75) |
Cimetidine was classified only as a weak inhibitor per the FDA Table of Inhibitors when determining concordance.
Concordance was defined as the inhibitor’s observed classification matched the classification per the FDA Table of Inhibitors.
Figure 2: AUC fold-change by inhibitor.
Observed AUC fold-change of the substrate is plotted by the inhibitor that caused the fold-change. Expected AUC fold-change is based on definitions by the FDA, where values of 1.25-<2 and ≥2-<5 translate to weak and moderate inhibitors respectively. The squares and diamonds represent observed AUC fold changes caused by moderate and weak inhibitors respectively. Each square or diamond represents one inhibitor-substrate pair and the total pairs per inhibitor is provided after the respective inhibitor. Cimetidine was classified as a weak inhibitor when determining concordance
Concordance of Inhibitor Classification in FDA Table of Inhibitors with the Product Labels
Among 17 drugs listed on the FDA Table of Inhibitors as a moderate or weak CYP2D6 inhibitor, PLs had the classification clearly stated (n=3), indicated that the drugs had unknown or did not possess clinically relevant inhibition (n=2), or lacked specific comments about enzyme inhibition of CYP2D6 (n=12). Of those with a classification clearly stated in the PL, two of the three inhibitors were concordant: duloxetine and mirabegron (moderate). While, cinacalcet is listed in the table as a moderate inhibitor, its PL states it is a strong inhibitor. The two inhibitors whose PL indicated unknown or no clinically relevant inhibition were escitalopram and desvenlafaxine, which are classified as weak inhibitors in the FDA Table of Inhibitors. Escitalopram’s PL states the clinical significance of CYP2D6 inhibition is unknown, while the label for desvenlafaxine states inhibition is minimal and not clinically relevant. Among the 11 PLs with AUC data (14 inhibitor-substrate pairs), eight pairs were concordant.
Phenotype Reporting
Of the 89 inhibitor-substrate pairs, 51 (57%) reported CYP2D6 phenotypes for the participants. The phenotype method varied greatly between pairs; 20 (39%) derived phenotype by genotyping alone, 20 (39%) used a probe drug alone (e.g., dextromethorphan, debrisoquine), while six (12%) used both genotyping and a probe drug, and for five (10%) the method was not reported. Some studies differed in how they grouped phenotypes for analysis. Among the 26 pairs that genotyped participants, 17 pairs reported data only in NMs, four grouped IMs and NMs together, three grouped NMs, IMs, and UMs together, one grouped NMs, IMs and PMs together, and one grouped NMs and UMs together.
Inhibitors with Conflicting Results
Of the 17 inhibitors included in all inhibitor-substrate pairs, five inhibitors (i.e., amiodarone, cobicistat, labetalol, lorcaserin, and vemurafenib) were excluded from the secondary outcome of evaluating additional study criteria as they only had one inhibitor-substrate pair. Of the five inhibitors excluded, all except lorcaserin were found to be concordant with their respective classification in the FDA Table of Inhibitors. Pairs of the remaining 12 inhibitors were evaluated. Table 2 describes the observed degree of inhibition and concordance by dose, substrate, and steady state. Table S3 contains the substrate doses. Six of the 12 inhibitors classified by the FDA Table of Inhibitors were observed to be concordant with their respective inhibition classification in most of the inhibitor-substrate pairs. Concordance was higher when inhibitors were at steady state for all 8 inhibitors that were studied at both steady state and pre-steady state. Similarly, overall concordance was numerically higher in pairs where the inhibitor was at steady state (72%) compared to overall (62%).
Table 2.
Secondary Outcome: Dose, Substrates, Steady-State
| Classification per FDA Table of Inhibitors |
Inhibitor Name | Observed inhibitor classification, n (%) |
Doses of Inhibitor studied, n (concordance %) |
Substrates studied, n (concordance %) |
Inhibitor Steady State Status, n (concordance %) |
|---|---|---|---|---|---|
| Moderate | Cinacalcet n = 4 |
Moderate, 3 (75) Insignificant, 1 (25) |
“25 or 100 mg”, 1 (0) 50 mg, 1 (100) 90 mg, 2 (100) |
Amitriptyline, 1 (100) Desipramine, 2 (100) Dextromethorphan, 1 (0) |
SS: 3, (100) Pre-SS: 1,(0) |
| Duloxetine, n = 9 |
Moderate, 4 (44) Weak, 4 (44) Insignificant, 1 (11) |
30 mg, 1 (0) 60 mg, 5 (40) 80 mg, 1 (0) 120 mg, 2 (100) |
Desipramine, 3 (100) Dextromethorphan, 2 (50) Metoprolol, 1 (0) Tolterodine, 1 (0) Tramadol, 2 (0) |
SS: 4 (50) Pre-SS: 4(25) Unknown: 1 (100) |
|
| Fluvoxamine, n = 7 |
Strong, 1 (14) Moderate, 2 (29) Weak, 3 (43) Insignificant, 1 (14) |
50 mg, 1 (0) 50-100 mga, 2 (0) 100 mg, 3 (67) 150 mg, 1 (0) |
Amitriptyline, 1 (0) Atomoxetine, 1 (0) Desipramine, 1 (0) Dextromethorphan, 1 (0) Imipramine, 2 (100) Nebivolol, 1 (0) |
SS: 6 (33) Pre-SS: 1 (0) |
|
| Mirabegron, n = 7 |
Moderate 5 (71) Weak, 1 (14) Insignificant, 1 (14) |
50 mg, 3 (33) 100 mg, 2 (100) 160 mg, 2 (100) |
Desipramine, 2 (100) Metoprolol, 2 (100) Tolterodine, 3 (33) |
SS: 2 (100) Pre-SS: 5 (40) |
|
| Weak | Abiraterone, n = 2 |
Moderate, 1 (50) Weak, 1 (50) |
1,000 mg, 2 (50) | Dextromethorphan, 2 (50) | SS: 1 (100) Unknown: 1 (0) |
| Celecoxib, n = 3 |
Weak, 1 (33) Insignificant, 2 (67) |
100 mg, 1 (0) 200 mg, 1 (0) 400 mg, 1 (100) |
Metoprolol, 1 (100) Tramadol, 2 (0) |
SS: 2 (50) Pre-SS: 1 (0) |
|
| Cimetidineb, n = 26 |
Moderate, 2 (8) Weak, 20 (77) Insignificant, 4 (15) |
400 mg, 2 (50) 600 mg, 1 (100) 800 mg, 8 (100) 1,000 mg, 6 (67) 1,200 mg, 9 (67) |
Amitriptyline, 1 (100) Desipramine, 2 (50) Imipramine, 3 (67) Metoprolol, 6 (83) Nebivolol, 1 (100) Nortriptyline, 1 (0) Propafenone, 2 (50) Propranolol, 9 (89) Venlafaxine, 1 (100) |
SS: 23, (78) Pre-SS: 3, (67) |
|
| Clobazam, n = 2 |
Weak, 2 (100) | 40 mg, 2 (100) | Dextromethorphan, 2 (100) | SS: 1 (100) Unknown: 1 (100) |
|
| Desvenlafaxine, n = 6 |
Weak, 3 (50) Insignificant, 3 (50) |
100 mg, 4 (50) 400 mg, 2 (100) |
Desipramine, 6 (50) | SS: 4 (50) Unknown: 2 (50) |
|
| Escitalopram, n = 3 |
Moderate, 1 (33) Weak, 2 (67) |
10 mg, 1 (100) 20 mg, 2 (50) |
Desipramine, 1 (0) Metoprolol, 1 (100) Tramadol, 1 (100) |
SS: 3 (67) | |
| Ritonavir, n = 5 |
Moderate, 3 (60) Weak, 2 (40) |
200 mg, 2 (100) 400 mg, 2 (0) 1,000 mg, 1 (0) |
Desipramine, 2 (50) Dextromethorphan, 3 (33) |
SS: 4 (50) Pre-SS: 1 (0) |
|
| Sertraline, n = 10 |
Moderate, 1 (10) Weak, 6 (60) Insignificant, 3 (30) |
50 mg, 5 (80) 100 mg, 1 (100) 150 mg, 4 (25) |
Desipramine, 6 (66) Imipramine, 2 (0) Metoprolol, 2 (100) |
SS: 6 (83) Pre-SS: 4 (25) |
50 mg daily for 3 days and 100 mg daily for 3 days
Cimetidine is also listed as a moderate inhibitor in the FDA Table of inhibitors, we only treated it as a weak inhibitor as the overall concordance with weak inhibitor was 77%, while it was 8% if treated as a moderate inhibitor.
The bolded classification within the column “observed inhibitor classification” represents the concordance with the classifications listed in FDA Table of Inhibitors.
SS: Steady state; Pre-SS: Pre steady-state
Table 3 describes data (e.g., multi-pathway drug interactions, genotype, phenotype) that may alter the role of CYP2D6 on the metabolism of a substrate. The strength of inhibition observed appeared to differ based on whether the drug inhibited multiple metabolic pathways for the substrate studied; this was specifically the case for cimetidine, fluvoxamine, and ritonavir.
Table 3.
Secondary Outcome: Multiple pathways, Phenotyping, and Genotyping
| Classification per FDA Table of Inhibitors |
Inhibitor | Inhibitor of additional pathways? |
CYP2D6 substrates studied with inhibitor metabolized by additional enzymes |
Phenotyping / Genotyping Mechanism, n |
Observed phenotype, n (concordance %) |
|---|---|---|---|---|---|
| Moderate | Cinacalcet | No | n/a | Genotyping, 1 Unknown, 3 |
NMs, 4 (75) |
| Duloxetine | No | n/a | Genotyping, 8 | NMs, 7 (29) Multiple (IMs, NMs, UMs), 1 (100) |
|
| Fluvoxamine | Inhibits CYP1A2, CYP2C9, CYP2C19, and CYP3A | Amitriptyline and imipramine: CYP2C19 dextromethorphan: CYP3A |
Multiple Probes, 4 Genotyping, 1 |
NMs, 4 (25) | |
| Multiple (NMs, PMs), 1 (0) | |||||
| Mirabegron | No | n/a | Genotyping, 5 | Multiple (IMs, NMs), 2 (0) | |
| NMs, 3 (10) | |||||
| Weak | Abiraterone | No | n/a | Unknown, 1 | NMs, 1 (100) |
| Celecoxib | No | n/a | Genotyping, 1 | NMs, 1 (100) | |
| Cimetidinea | Inhibits CYP1A2 and CYP3A | Amitriptyline and imipramine: CYP2D6, CYP1A2, and CYP2C19 Propranolol: CYP2D6 and CYP1A2 |
Debrisoquine probe, 5 | NM, 2 (50) | |
| PM, 1 (100) | |||||
| “Rapid hydroxylators”, 1 (100) | |||||
| “Slow hydroxylators”, 1 (0) | |||||
| Clobazam | No | n/a | 0 | n/a | |
| Desvenlafaxine | No | n/a | Genotyping, 4 | Multiple (IMs, NMs,), 2 (50) | |
| Multiple (IMs, NMs, PMs), 1 (100) | |||||
| Multiple (IMs, NMs, UMs), 1 (0) | |||||
| Escitalopram | No | n/a | Tramadol probe , 1 | NMs, 2 (100) | |
| Genotyping & Metoprolol probe, 1 | |||||
| Ritonavir | Inhibits CYP3A | Dextromethorphan: CYP2D6 and CYP3A | Dextromethorphan probe, 2 | NMs, 1 (100) | |
| Multiple (IMs, NMs, UMs), 1 (100) | |||||
| Genotyping, 2 | Multiple (IMs, NMs, PMs,), 2 (0) | ||||
| Sertraline | Inhibits CYP2C9 | n/a | Dextromethorphan probe, 8 | NMs, 10 (60) | |
| Genotyping, 2 |
n represents the number of inhibitor-substrate pairs and % represents concordance within the respective n.
Cimetidine is also listed as a moderate inhibitor in the FDA Table of Inhibitors, we only treated it as a weak inhibitor as the overall concordance with weak inhibitor was 77%, while it was 8% if treated as a moderate inhibitor.
Cimetidine displayed weak or insignificant inhibition in the 13 inhibitor-substrate pairs where the substrate was not metabolized by other enzymes also inhibited by cimetidine. The two instances where cimetidine exhibited moderate inhibition were when the substrate was known to be metabolized by CYP2D6 and additional pathways inhibited by cimetidine.
The four fluvoxamine-substrate pairs affected by more than one pathway exhibited weak (n=1), moderate (n=2), and strong inhibition (n=1). The pair where strong inhibition was observed used the lowest fluvoxamine dose, 50 mg daily. While, in the three inhibitor-substrate pairs where CYP2D6 was the only pathway inhibited by fluvoxamine for a substrate, fluvoxamine displayed only weak (n=2) or insignificant (n=1) inhibition.
For ritonavir, discordant results were when the substrate (i.e., dextromethorphan) was metabolized by additional enzymes that ritonavir inhibits (i.e., CYP3A) or if the dose was five times higher than the typical clinical dose of 200 mg/day. In these cases, ritonavir exhibited moderate inhibition (n=3).
Based on the assessments of concordance, and possible reasons for discordance with the current classification in the FDA Table of Inhibitors, we provide author interpretations in Table 4 for possible reasons for discordance. In Table 5 we provide recommendations for changes in the FDA Table of Inhibitors, along with caveats for selected drugs. Specifically, we recommend changes for: celecoxib, cimetidine, desvenlafaxine, and fluvoxamine.
Table 4.
Secondary Outcome: Author Interpretation of Potential Reasons for Findings of Inconsistency with the FDA table of Inhibitors
| Inhibitor | Classification per FDA Table of Inhibitors |
Author Interpretation |
|---|---|---|
| Abiraterone | Weak | Unclear mechanism for inconsistent results |
| Celecoxib | Weak | Total daily doses ≤ 200 mg did not exhibit inhibition while 400 mg exhibited weak inhibition |
| Cimetidine | Weak & Moderate | Typically a weak inhibitor; only exhibited moderate inhibition with substrates affected by multiple pathways inhibited by cimetidine |
| Cinacalcet | Moderate | Typically a moderate inhibitor; multiple possibilities for inconsistent results (i.e., dose, steady state, and substrate sensitivity) |
| Clobazam | Weak | Always displayed weak inhibition although it was only studied with one substrate and one dosing strategy |
| Desvenlafaxine | Weak | Insignificant inhibition at clinically relevant doses |
| Duloxetine | Moderate | Unclear mechanism for inconsistent results |
| Escitalopram | Weak | All published evidence assessed supports the FDA classification as a weak inhibitor, although the FDA product label reports an AUC-fold increase that is at the lower limit of moderate inhibition |
| Fluvoxamine | Moderate | Weak inhibitor unless the substrate is affected by multiple pathways inhibited by fluvoxamine |
| Mirabegron | Moderate | Labeled dose exhibits moderate inhibition in NMs, but weak inhibition in populations that include IMs. Daily doses above the product labeling regardless of phenotype exhibit moderate inhibition. |
| Ritonavir | Weak | Weak inhibitor at typical doses unless the substrate is affected by multiple pathways inhibited by ritonavir |
| Sertraline | Weak | Weak inhibitor once sertraline reaches steady state |
Concordance was defined as the inhibitor’s observed classification matched the classification per the FDA Table of Inhibitors.
Table 5:
Recommendations for FDA Table of Inhibitors
| Moderate Inhibitor | Weak Inhibitor | Removal from table |
|---|---|---|
| cimetidine, cinacalcet, duloxetine, fluvoxamine, mirabegrona | abiraterone, amiodarone, celecoxibb, cimetidine, clobazam, cobicistat, desvenlafaxine, escitalopram, fluvoxamine, labetalol, lorcaserin, ritonavir, sertraline, vemurafenib | Desvenlafaxine |
Genotype should be considered, appears to moderately inhibit only when individuals are NMs
Only at doses > 200 mg total daily dose
DISCUSSION
This scoping review identified 60 records from publicly available literature describing AUC-fold change from moderate or weak CYP2D6 inhibitors. This resulted in 89 inhibitor-substrate pairs where 62% of the observed inhibitor classifications were concordant with the classifications per the FDA Table of Inhibitors. Study design may contribute to discordance and therefore our secondary outcome evaluated relevant study criteria. These results led to recommendations to modify the classifications in the FDA Table of Inhibitors for: celecoxib, cimetidine, desvenlafaxine, and fluvoxamine (Table 5). The rationale for these recommendations is discussed below.
Desvenlafaxine appeared to exhibit dose-related CYP2D6 inhibition as 400 mg dosing consistently exhibited weak inhibition (reported mean AUC fold-changes: 1.83 and 1.9) while 100 mg dosing exhibited insignificant inhibitory effects (maximum AUC fold-change: 1.26). Given these data and that all available studies examined doses above FDA-labeled dosing (i.e., 50mg), it seems reasonable to remove desvenlafaxine from the FDA Table of Inhibitors. Similarly, celecoxib was only observed to exhibit weak inhibition at higher doses, 400 mg daily, typically reserved for select conditions (e.g., rheumatoid arthritis, ankylosing spondylitis). Given the FDA label permits this dosing in select conditions, we agree it is appropriate to classify celecoxib as a weak inhibitor. However, we recommend adding that lower doses (e.g., 200 mg daily), as indicated for osteoarthritis, have not exhibited CYP2D6 inhibition.
Cimetidine rarely exhibited moderate inhibition (2 of 26 inhibitor-substrate pairs; 8%). All observations of moderate inhibition occurred when studied with a substrate that is also metabolized by CYP1A2 (i.e., imipramine, propranolol), which cimetidine also inhibits. The dual pathway inhibition can explain the higher AUC-fold change and, if unaccounted for, overstate the inhibitory effect of cimetidine on CYP2D6. Therefore, the classification per the FDA Table of Inhibitors overstates the inhibition effect of cimetidine by classifying it as moderate. Fluvoxamine was similar to cimetidine; in instances where fluvoxamine exhibited moderate or strong inhibition, the substrate was metabolized by at least two pathways that fluvoxamine inhibits. In pairs not confounded by multiple pathways, both fluvoxamine and cimetidine generally exhibited weak inhibition and therefore, should be reclassified as weak inhibitors.
The literature broadly supports sertraline’s classification as a weak inhibitor when limited to inhibitor-substrate pairs where sertraline was at steady state (5 of 6 inhibitor-substrate pairs; 83%) but not at pre-steady state (1 of 4 inhibitor-substrate pairs; 25%). Interestingly, the presence of steady state appeared more important than dose, while the PL indicates dose-related inhibition. Thus, studies in which sertraline was used in a clinically relevant manner (steady state), the data are consistent with the classification of weak inhibitor.
Drug-Drug-Gene Interactions
The concept of a DDI is analogous to a drug-gene interaction (DGI), and a DDGI is when both are taken into consideration.(18) In a pharmacokinetic sense, regardless if it is a lack of enzyme production (DGI) or drug-mediated inhibition of an enzyme (DDI), the result is reduced metabolism of the substrate. A DDGI incorporates both drug and genotype into the assessment; otherwise CYP2D6 genotype may act as a confounder in DDI studies if unaccounted for (similar to drug-interactions confounding pharmacogenetic studies). Unlike most of the medications classified as CYP2D6 inhibitors, two more recently approved drugs classified as CYP2D6 inhibitors, duloxetine and mirabegron, often reported genotype data in their respective records.
Duloxetine records generated inhibitor-substrate pairs with observed moderate (n=4) and weak (n=4) inhibition classifications. Conflicting factors (e.g., dose, genotype, substrate) make it difficult to define an inhibitory classification for duloxetine. For example, the article by Storelli et. al. produced 4 inhibitor-substrate pairs included in this review. The authors examined CYP2D6 activity score, which is a way to quantify enzyme activity. They examined the effect of duloxetine on two different substrates (i.e., tramadol, dextromethorphan) across different CYP2D6 activities (i.e., activity score=1, activity score=2).(6) The activity score of two (i.e., more enzyme activity) exhibited higher AUC fold-change than an activity score of one with dextromethorphan (2.4 vs 1.8, respectively) and tramadol (1.7 vs 1.2, respectively). Thus, duloxetine caused a higher AUC-fold change when subjects possessed higher enzyme activity. Additionally, dextromethorphan and tramadol were administered concomitantly, which may have resulted in competitive inhibition of CYP2D6. Specifically, duloxetine was observed to be a moderate inhibitor when examining AUC for dextromethorphan but not for tramadol. As dextromethorphan has a higher affinity for CYP2D6 than tramadol,(19) it may have blunted the inhibitory effect of duloxetine on tramadol. Thus, defining the inhibitor classification of duloxetine was limited by two major confounding factors (i.e., CYP2D6 genotype and competitive inhibition).
In addition to data from Storelli et. al. on duloxetine, others have previously explored the relationship between magnitude of enzyme activity and magnitude of CYP2D6 inhibitory effects. Tod et. al. recognized the fraction of clearance for a substrate increases with enzyme activity (i.e., PM < IM < NM < UM) likewise, the magnitude of the AUC fold-change from a DDI increases with enzyme activity (i.e., PM < IM < NM < UM).(20, 21) We would not expect any AUC fold-change in PMs as a result of a DDI. An example of this concept is seen with the data describing mirabegron’s CYP2D6 inhibitory properties. Mirabegron exhibited moderate inhibition in all instances where assessments were restricted to CYP2D6 NMs (3 of 3 inhibitor-substrate pairs). The degree of inhibition was reduced to weak, or blunted as evidenced by a lower AUC ratio, when the population included NMs and IMs (2 of 2 inhibitor-substrate pairs). In other words, a moderate or weak CYP2D6 inhibitor may exhibit blunted CYP2D6 inhibitory pharmacokinetic properties in patients with reduced CYP2D6 activity. Higher CYP2D6 activity (inferred by genotype) resulted increased inhibitory effects (i.e., higher AUC-fold change) for mirabegron and duloxetine, which highlights the importance of collecting genotype data to accurately assess inhibitory properties. Although IMs have a lower observed AUC ratio than NMs, the inhibitor still causes a clinically significant interaction. IMs likely have higher AUCs than NMs in the absence of an inhibitor, thus even with the same fold-elevation of drug concentration between IM and NM in the presence of an inhibitor, the IMs would likely have higher absolute drug concentrations.
It is unfortunate that only 26 of 89 (29%) inhibitor-substrate pairs reported CYP2D6 genotype, however it is not unexpected given that many studies were published before pharmacogenetic effects were well recognized and routinely considered in clinical pharmacokinetic studies. Unless accounting for genotype, studies may be confounded by the assumption that patients exhibit normal metabolism at baseline.(21) Recently, the FDA issued a draft guidance for clinical drug interaction studies, which recommended DNA collection from subjects “to characterize differences in the magnitude of the DDI across genotype groups and to understand why some subjects have unusual increases or decreases in drug concentrations.”(22) To maximize this benefit in future studies, investigators should consider analyzing genotype-inferred phenotype groups in a meaningful way. No studies compared AUC fold-changes between different phenotype groups, which limited our ability to understand how CYP2D6 inhibitors are affected by genotypic inter-individual variability.
Additionally, the lack of genotype data in studies limits extrapolation to comprehensive predictions of CYP2D6 function (i.e., clinical phenotype), which are needed when implementing pharmacogenetics Some approaches treat moderate and weak inhibitors similarly (e.g., reduce CYP2D6 activity score by half)(23) while others ignore weak inhibitors.(7, 8) More data are needed in this area, but it seems plausible that weak inhibitors can be ignored, barring specific circumstances (e.g., substrate has a narrow therapeutic index, multiple pathways affected, multiple drugs affecting CYP2D6).
FDA Product Label
In comparing different FDA resources, the FDA PLs and FDA Table of Clinical Inhibitors were notably inconsistent. Out of the 17 drugs listed as a moderate or weak inhibitor in the FDA Table of Clinical Inhibitors, the FDA PL matched in just two, duloxetine and mirabegron. This lack of consistency may be influenced by the date of FDA approvals or outdated updates.
Strengths, Limitations, and Conclusions
To our knowledge, this study is the first to assess whether the classifications of inhibitors listed as moderate and weak in the FDA Table of Inhibitors is supported by publicly available primary literature. This work has resulted in recommended changes for the FDA Table of Inhibitors. Strengths of this study include the quantity and quality of clinical data supporting our recommendations compared to the references in the FDA Table of Clinical Inhibitors.(12) Secondly, the secondary outcome evaluated factors that may influence inhibitor-substrate pairs, which allowed us to identify clinically-relevant factors that appear to affect the magnitude of CYP2D6 inhibition. Importantly, this included CYP2D6 genotype, when available, and the assessment of substrates affected by multiple pathways influenced by the suspected CYP2D6 inhibitor.
Limitations to this study include that it is a scoping review, which did not account for unpublished literature, New Drug Applications, and was limited to articles published in English. The literature search may not have captured all relevant data. . This study was also limited by widespread confounders (e.g., other pharmacokinetic pathways, conduct of the study when the inhibitor was not at steady state, CYP2D6 genotype/phenotype, lack of standardized CYP2D6 genotype translation) within the articles assessed. The FDA draft guidance indicates that when studying DDIs, a single dose of an inhibitor may be used to extrapolate to steady-state concentrations when administered with a substrate that exhibits linear kinetics. If substrates have non-linear pharmacokinetics then both the substrate and inhibitor should be at steady state. As studies may have been conducted prior to the FDA draft guidance, the studies may not align with the guidance.(22) Likewise, studies did not assess genotypes for enzymes that metabolize inhibitors (e.g., CYP2C19 poor metabolism is associated with increased escitalopram concentrations, which may increase the magnitude of CYP2D6 inhibition). Furthermore, observed inhibitor classifications are based on AUC-fold change threshold definitions from the FDA, which are defined for sensitive substrates per the FDA Table of Substrates. There are no defined AUC fold-change thresholds when inhibitors are given with moderate-sensitive substrates, thus we extrapolated the definition. As seen in Table 1, concordance did not appear to vary based on the classification of substrate. Data obtained from FDA PLs were limited as they may lack many of the desired details, including references. Therefore, it is possible that a publication and information in the FDA PL are using the same data, and thus duplicated in our results.
The FDA table of Inhibitors is not “intended to be an exhaustive list” of CYP2D6 inhibitors, however, this is a problem that extends beyond CYP2D6 to all drug-drug interactions.(12) There is a clear clinical need for a gold standard reference of inhibitors as drug interactions may affect many patients. Current DDI tools are markedly inconsistent.(11) Enhanced attention by the FDA to this area may be one way to start. However, it is not feasible to study every drug-drug pair in humans, let alone every drug-drug-gene pair. Innovative solutions such as physiologically-based pharmacokinetic modeling (PBPK) have been proven valuable within the drug-drug interaction field and may be a solution to evaluate gene-drug-interactions too.(24-28)
In summary, we suggest the FDA modify the CYP2D6 inhibitor classifications for desvenlafaxine (remove), cimetidine (downgrade to weak inhibitor), fluvoxamine (downgrade to weak inhibitor) and to make notations to mirabegron, and celecoxibas described in Table 5. More DDI studies are needed to assess additional inhibitor-substrate pairs, explore the combinatorial effects of genotype and drug interaction data, and assess the clinical impact of these interactions.
Supplementary Material
Table S1: FDA Examples of Clinical Inhibitors and Clinical Substrates for CYP2D6
Table S2: All Inhibitor-Substrate Pairs and Respective Record Reference
Table S3: Concordance by Substrate
STUDY HIGHLIGHTS.
- What is the current knowledge on the topic?
- CYP2D6 inhibitors classified by the FDA as strong lead to null CYP2D6 enzyme activity, while it is less clear with moderate or weak inhibitors because of uncertainty in the magnitude of CYP2D6 inhibition caused.
- What question did this study address?
- Do the FDA-defined thresholds of AUC-fold increase of substrates result in moderate and weak inhibitor classifications of CYP2D6 inhibitors found in the FDA Table of Inhibitors?
- What does this study add to our knowledge?
- The FDA definitions of CYP2D6 inhibitors may not be able to broadly be applied to every drug, there may be specific details for inhibitors that we should be aware of when applying to drug interaction data to infer a CYP2D6 phenotype..
- How might this change clinical pharmacology or translational science?
- We propose that the FDA modify the FDA Table of Inhibitors by re-classifying certain drugs and adding clarifications to make clinicians aware of drug-specific details that need to be considered before broadly quantifying the strength of CYP2D6 inhibition and applying to practice.
ACKNOWLEDGEMENTS
Funding: Work is supported by NIH/NHGRI (U01HG 007269) [LHC, JAJ] and NIH/NCATS (UL1TR001427) [LHC].
As an Editor-in-Training for Clinical Pharmacology & Therapeutics, Emily Cicali was not involved in the review or decision process for this paper.
Footnotes
Conflict of Interest Statement: All authors declared no competing interests for this work
REFERENCES
- (1).Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE & Zeldin DC The human intestinal cytochrome P450 “pie”. Drug metabolism and disposition: the biological fate of chemicals 34, 880–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wilkinson GR Drug metabolism and variability among patients in drug response. N Engl J Med 352, 2211–21 (2005). [DOI] [PubMed] [Google Scholar]
- (3).Evans WE & McLeod HL Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med 348, 538–49 (2003). [DOI] [PubMed] [Google Scholar]
- (4).Caudle KE et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genetics in medicine : official journal of the American College of Medical Genetics 19, 215–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Verbeurgt P, Mamiya T & Oesterheld J How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics 15, 655–65 (2014). [DOI] [PubMed] [Google Scholar]
- (6).Storelli F, Matthey A, Lenglet S, Thomas A, Desmeules J & Daali Y Impact of CYP2D6 Functional Allelic Variations on Phenoconversion and Drug-Drug Interactions. Clinical pharmacology and therapeutics 104, 148–57 (2018). [DOI] [PubMed] [Google Scholar]
- (7).Knisely MR et al. CYP2D6 drug-gene and drug-drug-gene interactions among patients prescribed pharmacogenetically actionable opioids. Applied nursing research : ANR 38, 107–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Smith D et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genetics in medicine: official journal of the American College of Medical Genetics 21, 1842–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cicali EJ et al. Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene-drug pairs across ambulatory care settings. Genetics in medicine: official journal of the American College of Medical Genetics 21, 2264–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cavallari LH et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genetics in medicine: official journal of the American College of Medical Genetics 21, 2255–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Patel RI & Beckett RD Evaluation of resources for analyzing drug interactions. J Med Libr Assoc 104, 290–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).U.S. Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labeling. Accessed 01/10/2019 <https://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm093664.htm> (2019).
- (13).Flockhart. The Flockhart Table™. <https://drug-interactions.medicine.iu.edu/Main-Table.aspx> (2018).
- (14).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clinical pharmacology and therapeutics 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Frost DA, Soric MM, Kaiser R & Neugebauer RE Efficacy of Tramadol for Pain Management in Patients Receiving Strong Cytochrome P450 2D6 Inhibitors. Pharmacotherapy 39, 724–9 (2019). [DOI] [PubMed] [Google Scholar]
- (16).Ouzzani M, Hammady H, Fedorowicz Z & Elmagarmid A Rayyan-a web and mobile app for systematic reviews. Syst Rev 5, 210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Clinical Pharmacogenetics Implementation Consortium. Accessed 01/10/2019 <https://cpicpgx.org/> (2019).
- (18).Conrado DJ, Rogers HL, Zineh I & Pacanowski MA Consistency of drug-drug and gene-drug interaction information in US FDA-approved drug labels. Pharmacogenomics 14, 215–23 (2013). [DOI] [PubMed] [Google Scholar]
- (19).Frank D, Jaehde U & Fuhr U Evaluation of probe drugs and pharmacokinetic metrics for CYP2D6 phenotyping. European journal of clinical pharmacology 63, 321–33 (2007). [DOI] [PubMed] [Google Scholar]
- (20).Tod M, Goutelle S, Clavel-Grabit F, Nicolas G & Charpiat B Quantitative prediction of cytochrome P450 (CYP) 2D6-mediated drug interactions. Clinical pharmacokinetics 50, 519–30 (2011). [DOI] [PubMed] [Google Scholar]
- (21).Fermier N, Bourguignon L, Goutelle S, Bleyzac N & Tod M Identification of Cytochrome P450-Mediated Drug-Drug Interactions at Risk in Cases of Gene Polymorphisms by Using a Quantitative Prediction Model. Clinical pharmacokinetics 57, 1581–91 (2018). [DOI] [PubMed] [Google Scholar]
- (22).Clinical Drug Interaction Studies — Study Design, Data Analysis, and Clinical Implications Guidance for Industry. Accessed 3/5/19 <https://www.fda.gov/downloads/drugs/guidances/ucm292362.pdf> (2017).
- (23).Borges S et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. Journal of clinical pharmacology 50, 450–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jamei M Recent Advances in Development and Application of Physiologically-Based Pharmacokinetic (PBPK) Models: a Transition from Academic Curiosity to Regulatory Acceptance. Current pharmacology reports 2, 161–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rowland M, Lesko LJ & Rostami-Hodjegan A Physiologically Based Pharmacokinetics Is Impacting Drug Development and Regulatory Decision Making. CPT: pharmacometrics & systems pharmacology 4, 313–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lagishetty CV, Deng J, Lesko LJ, Rogers H, Pacanowski M & Schmidt S How Informative Are Drug-Drug Interactions of Gene-Drug Interactions? Journal of clinical pharmacology 56, 1221–31 (2016). [DOI] [PubMed] [Google Scholar]
- (27).Gong J, Iacono L, Iyer RA, Humphreys WG & Zheng M Physiologically-based pharmacokinetic modelling of a CYP2C19 substrate, BMS-823778, utilizing pharmacogenetic data. British journal of clinical pharmacology 84, 1335–45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Storelli F, Desmeules J & Daali Y Physiologically-Based Pharmacokinetic Modeling for the Prediction of CYP2D6-Mediated Gene-Drug-Drug Interactions. CPT: pharmacometrics & systems pharmacology 8, 567–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: FDA Examples of Clinical Inhibitors and Clinical Substrates for CYP2D6
Table S2: All Inhibitor-Substrate Pairs and Respective Record Reference
Table S3: Concordance by Substrate