Figure 2.
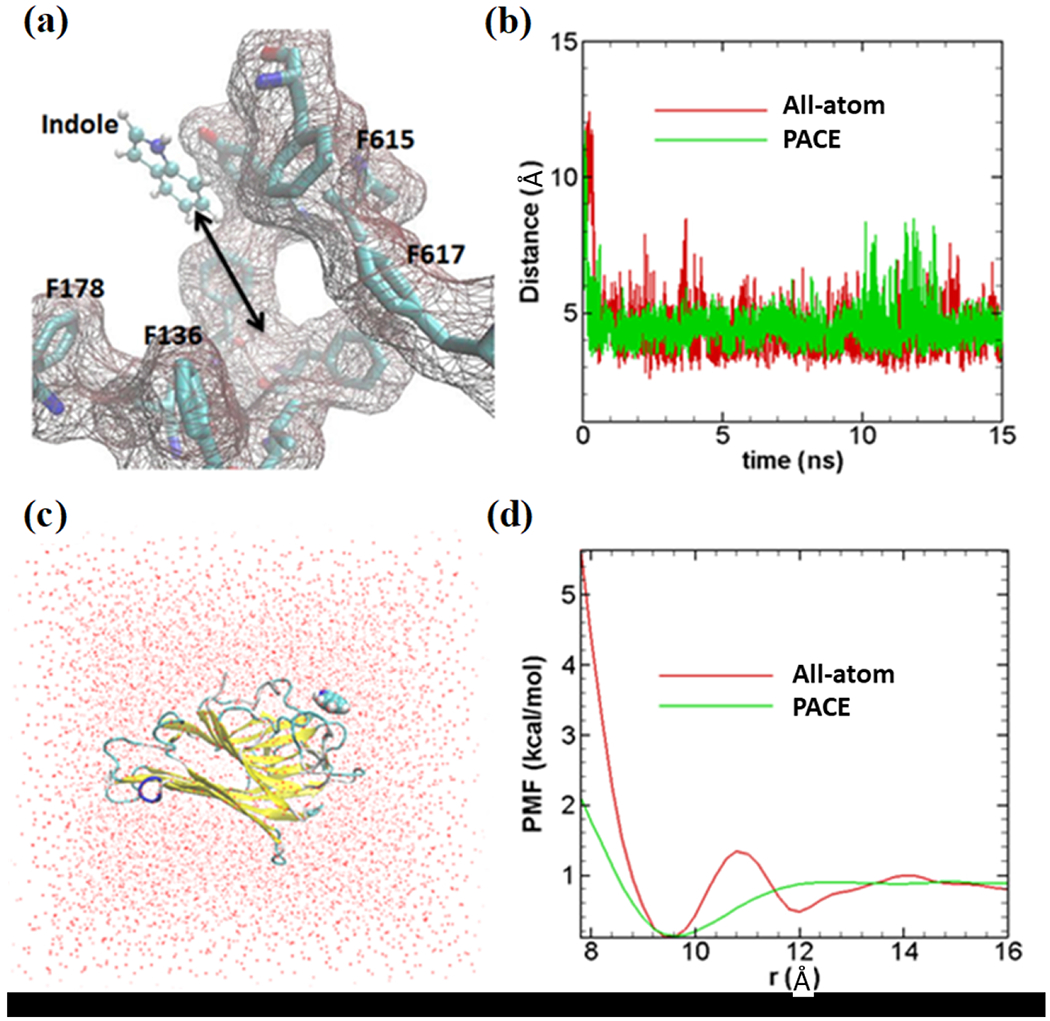
Validation of coarse-grained indole force field. (a) All-atom model of indole molecule has been shown in ball and stick representation; binding site with important amino acids are also shown. (b) Distances between indole center of mass and center of 4 amino acids shown in Figure 2a. All-atom indole binds to binding site very quickly (within ~1 ns) and stays close to the binding site as presented with a red line. United-atom indole (green line) shows similar dynamics as all-atom simulation. (c) All-atom simulation model (periodic box of 48×48×48 Å3) to calculate PMF; red dots are representing water molecules. Indole binding protein (PDB ID: 3SNM) is shown in new-cartoon representation. (d) PMF profiles for both methods. All-atom PMF, indicated by red lines, (~1kcal/mol) matches fairly well with PACE PMF (green line). Indole has been constrained to move unidirectionally while calculating PMF both in all-atom and PACE simulations.
