Figure 5. Validation of a duplexed qPCR assay for quantifying Eggerthella lenta and specific genes of interest.
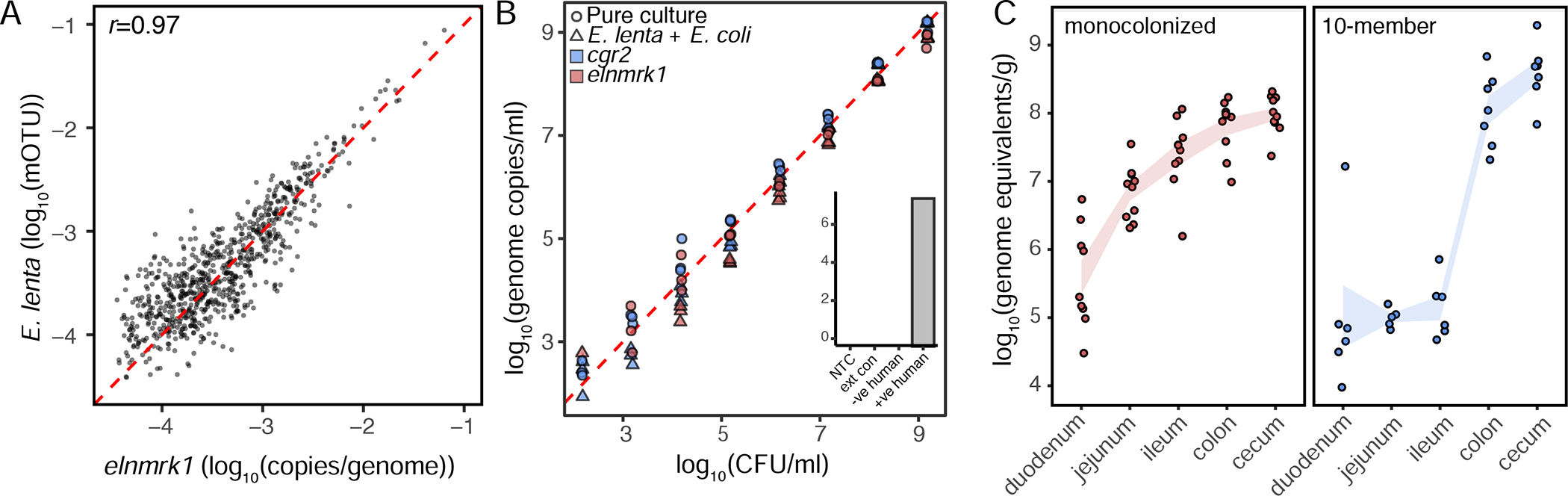
(A) Correlation of elnmrk1 and E. lenta abundances in metagenomes reveals a high degree of correlation (p<0.001, r=0.97) and little evidence of an elnmrk1-negative E. lenta population in 760 human GI tracts (red dashed line x=y). (B) Validation of assay sensitivity demonstrates a practical quantification limit of 1400 genome copies and that the presence of a second more abundant, and easily lysed, organism (4E9 CFU/mL E. coli K12 MG1655) does not prevent the recovery and quantification of E. lenta (inset controls: no-template control (NTC), blank DNA extraction control (ext con), and negative and positive human controls. (C) Quantification of E. lenta DSM 2243 in gnotobiotic mice monocolonized or colonized with a 7-member synthetic community. E. lenta is detectable along the entire length of the gastrointestinal tract with highest levels observed in the colon and cecum. Points represent individual animals (n=6–10/group) and lines represent mean±sem.
