Figure 1. RB-TnSeq screen to identify Salmonella genes involved in growth and colonization of the murine gut.
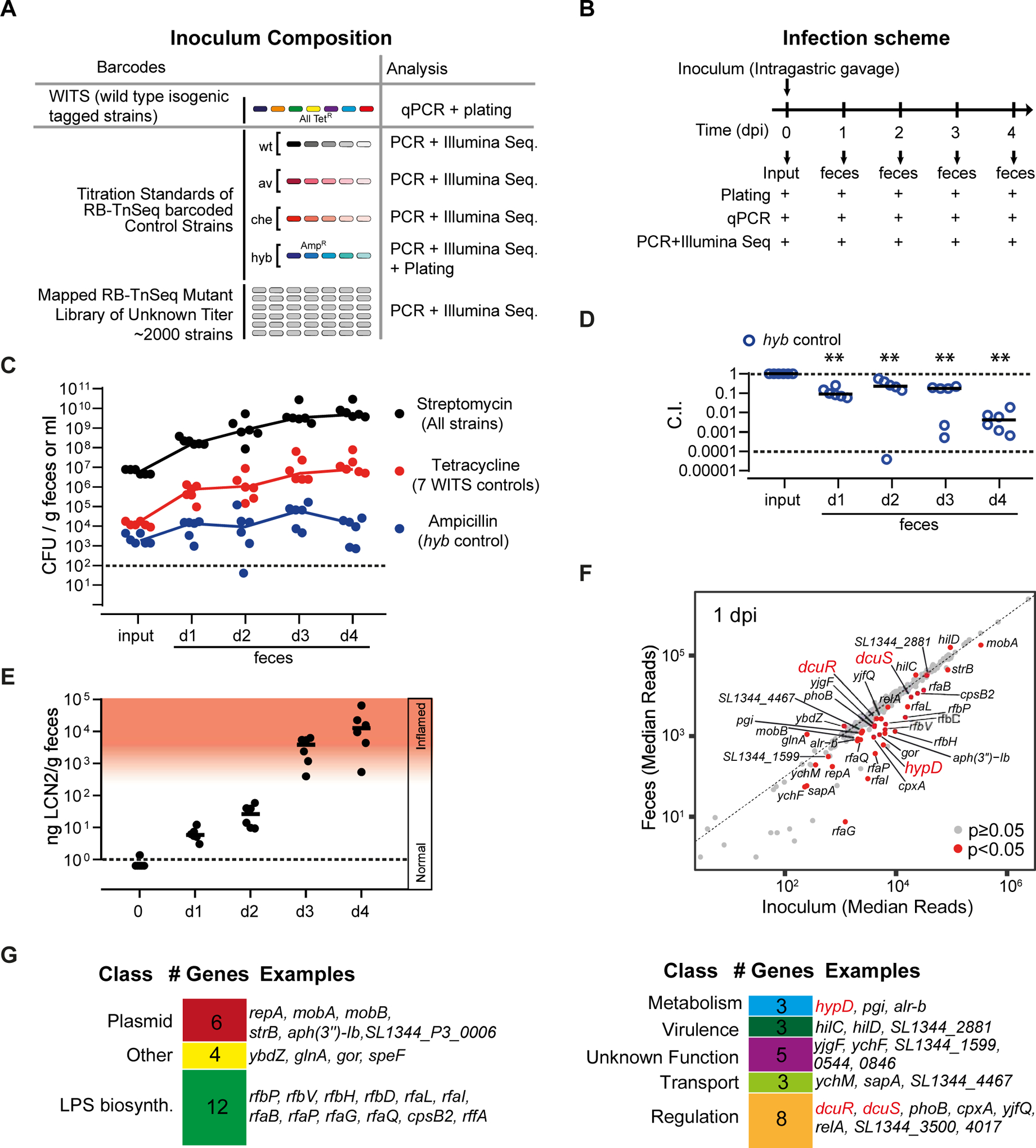
A. Composition of the inoculum pool. WITS-barcoded wild type controls were spiked in at densities of 1/2000 and titration standards of RB-TnSeq barcoded wild type and mutant controls as detailed in Star Methods and the Key Resource Table. B. Mouse infection protocol and sampling procedures. C. S. Typhimurium loads in the inoculum and the feces at days 1–4 pi as determined by plating. Black: Total S. Typhimurium loads. Red: total loads of WITS-barcoded wild type controls. Blue: Loads of the hyb mutant. Dotted line: detection limit. D. Attenuation of the hyb control strain (C.I.). E. Gut inflammation as measured by Lipocalin-2 ELISA. F. Fitness defects of RB-TnSeq mutants. We compared the relative barcode read abundances from the inoculum and the feces (day 1 pi; full data shown in Tab. S3). Red dots: mutants with significantly reduced or enhanced fitness compared to the wild type control strains (p < 0.05). G. Gene classes represented among the 43 RB-TnSeq hits implicated in initial growth.
