Figure 4. Aspartate and malate are important nutrients used to fuel anaerobic respiration.
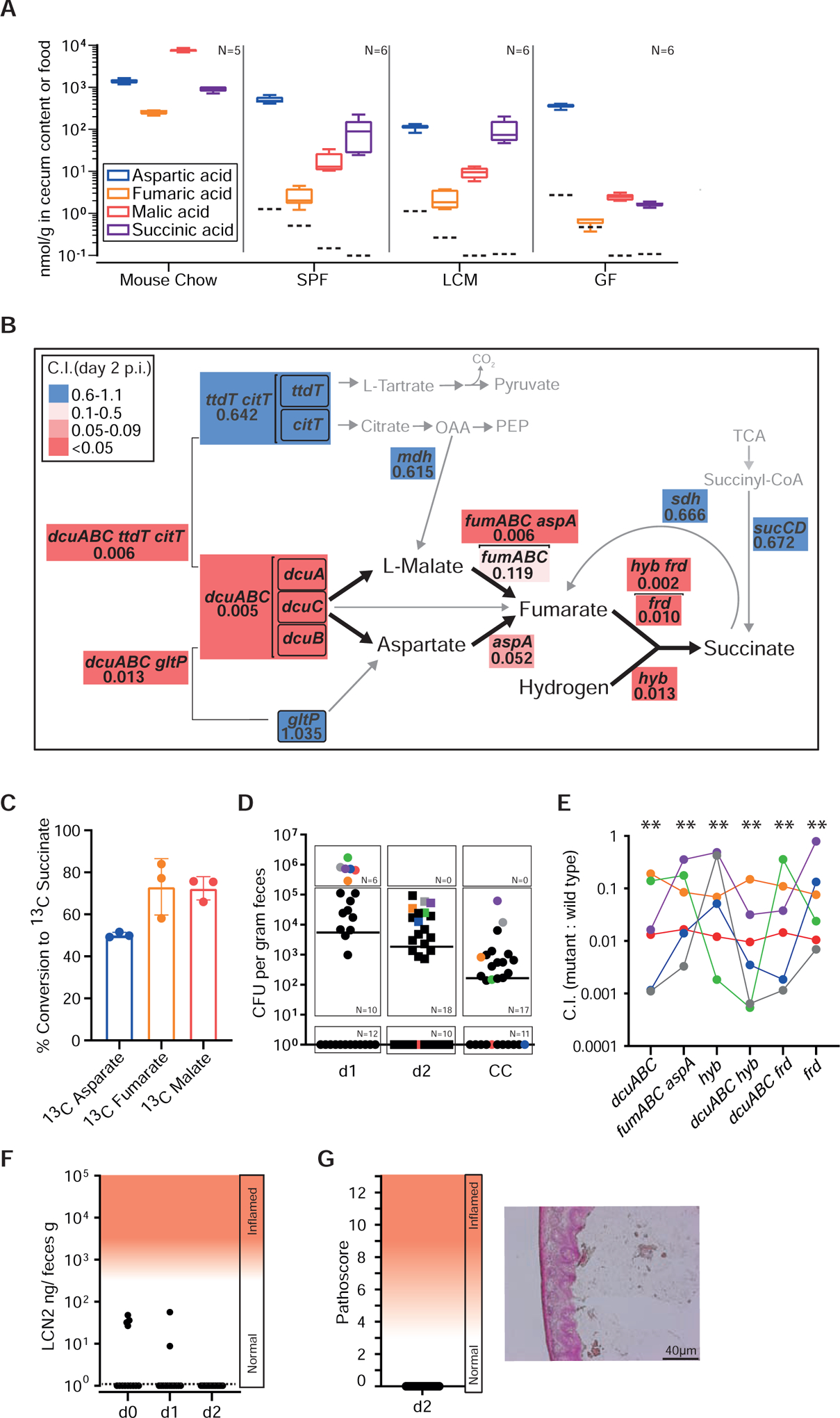
A. C4-Dicarboxlyate levels in the cecum lumen. Cecum contents (without particulate matter or cells) was recovered from mouse chow, germ free (GF), specific pathogen free (CONE), and LCM mice and analyzed by liquid chromatography mass spectrometry (LCMS). Independent measurements of n = 5 (mouse chow) and n = 6 (cecum samples) biological replicates. Dashed lines indicate the limits of detection; box plots extend from the 25th to 75th percentiles with whiskers going down to the smallest value and up to the largest. Center lines indicate median values. B. Competitive indices of site-directed mutants vs. wild type S. Typhimurium. We infected LCM mice with WITS-barcoded S. Typhimurium strain mixtures (5×106 cfu in total, by gavage; at least 6 independent experiments for each mutant) and analyzed the C.I. at days 1–4 pi by qPCR (Materials and Methods). All data are summarized in Tab. S3. Panel B shows data for day 2 pi. Gene symbols denote the mutants, analyzed. Arrows: metabolic conversions; Brackets indicate pathways that were collectively deleted. Numbers: median C.I. at day 2 pi; Colored boxes indicate the degree of attenuation, as indicated in the legend. Grey: Genes that are not essential for C4-dicarboxylate driven H2/fumarate respiration (mutant C.I. values 0.6–1.1). C. Conversion of 13C C4-dicarboxylates by S. Typhimurium. Wild type S. Typhimurium was grown anaerobically (37°C, 4% H2, 10% CO2, 86% N2) in minimal medium containing 45 mM 12C pyruvate and 10 mM of 13C aspartate, 13C fumarate or 13C L-malate (Star Methods). After reaching stationary phase, cell free culture supernatants were harvested and the amounts of released 13C succinate were analyzed by LC-MS (n = 3 independent cultures). D-G. Competitive infection experiment in CONE mice. N = 30 animals were infected for two days with wild type S. Typhimurium, S.TmdcuABC, S.TmfumAC fumB aspA, S.Tmhyb, S.TmdcuABC hyb, S.TmdcuABC frd, or S.Tmfrd (1:1:1:1:1:1:1 mix; 5×107 cfu in total, by gavage). D. Total fecal Salmonella loads as determined by plating. E. Competitive index, as determined by qPCR analysis of the strains’ barcodes. F. Gut inflammation as measured in feces by Lipocalin-2 ELISA. G. Histopathology of the infected cecum tissue at day 2 pi.
