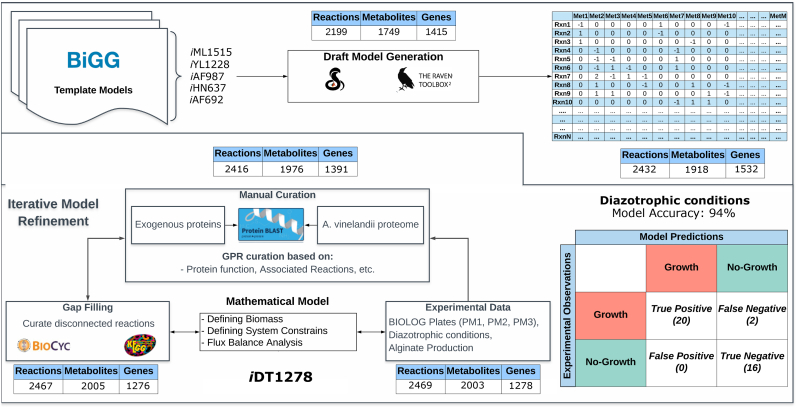
Fig. 1.
Workflow used to reconstruct a metabolic model of A. vinelandii DJ. A draft model was created from five template models present in BiGG (Escherichia coli str. K-12 substr. MG1655, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Geobacter metallireducens GS-15, Clostridium ljungdahlii DSM 13528 and Methanosarcina barkeri str. Fusaro). The RAVEN toolbox Agren et al., 2013 for MATLAB was used to create the draft model from stoichiometric data. The initial draft model contained 2,432 reactions, 1,918 metabolites, and 1,532 genes. The iterative process of model refinement included manual curation, gap-filling and curation using experimental data. The resultant final model contained A. vinelandii specific metabolic processes such as nitrogen fixation, and production of alginate and PHB. The final model, containing 2,469 reactions, 2,003 metabolites, and 1,278 genes, predicted with 94% accuracy.