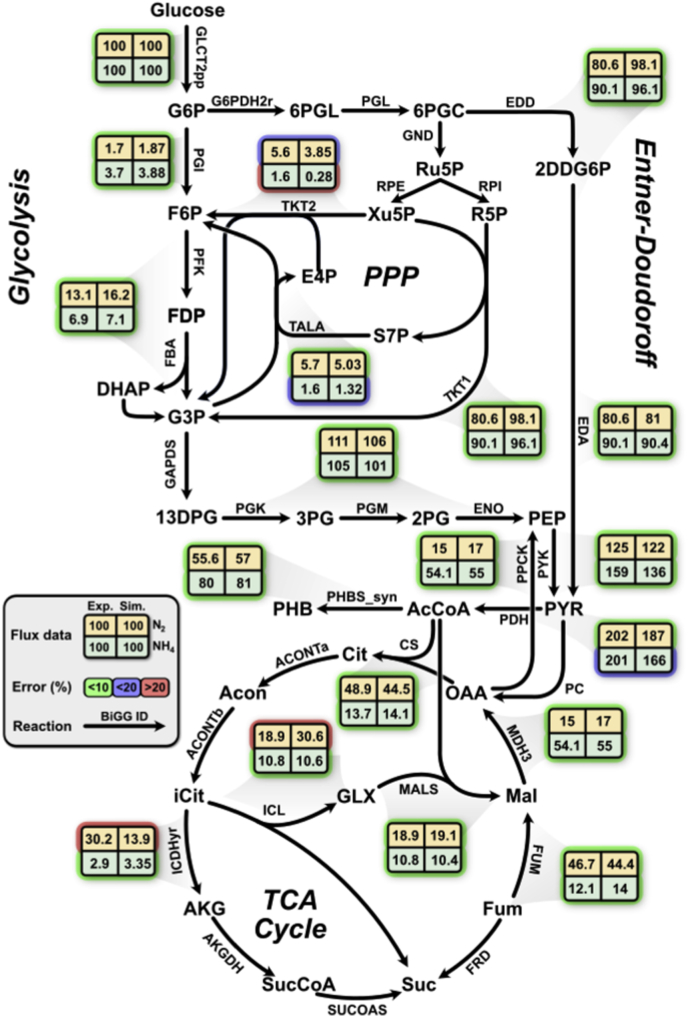
Fig. 5.
Metabolic flux map distribution of A. vinelandii under diazotrophic and non-diazotrophic conditions. The map displays major metabolic pathways involved in the PHB production. Values of metabolic flux are normalized to a glucose uptake rate of 100. Predicted metabolic fluxes were compared against fluxomic data determined by Wu et al., in 2019. The reactions were labeled according to their percent error (green, blue and red) and nitrogen source (green and yellow). Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FDP, fructose-1,6-bisphosphate; G3P, 3-phosphoglycerate; DHAP, dihydroxyacetone phosphate; 13DPG, 3-phosphoglyceroil phosphate; 2 PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; Cit, citrate; Acon, aconitate; iCit, isocitrate; AKG, a-ketoglutarate; sucCoA, succinyl coenzyme A; Suc, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; GLX, glyoxylate; PHB, polyhydroxybutyrate; 6PGL, 6-phospho-glucono-1,5-lactone; 6PGC, 6-phospho-gluconate; 2DDG6P, 2-Dehydro-3-deoxygluconate 6-phosphate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)