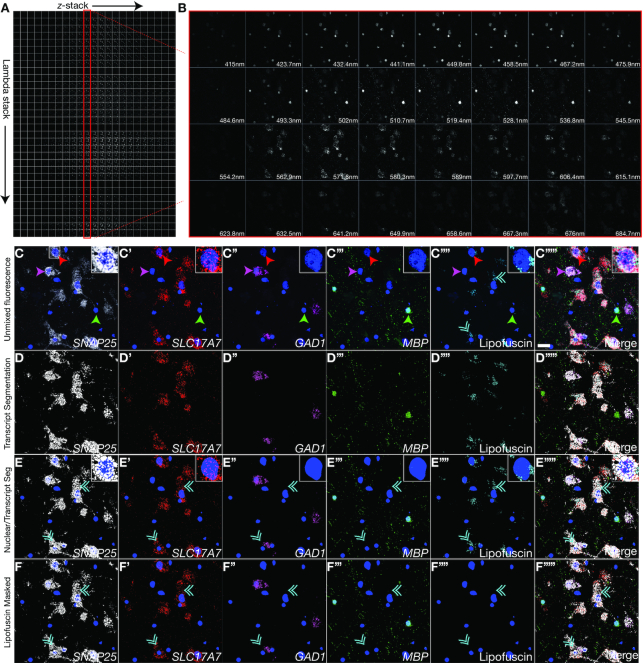
Figure 4.
Visualization and quantification of single transcripts in post-mortem human brain tissue using spectral imaging/linear unmixing and dotdotdot. (A) Matrix of raw confocal images acquired during spectral imaging in z-series of a single field of post-mortem human cortex at 63x magnification. A lambda stack is captured at each z-plane detecting expression of SNAP25, SLC17A7, GAD1, MBP (labeled using Opal690 [emission maximum at 690nm], Opal570 [emission maximum at 570 nm], Opal620 [emission maximum at 620 nm], and Opal 520 [emission maximum at 520 nm] dyes, respectively), and lipofuscin autofluorescence. (B) Representative lambda stack depicting a single z-plane acquired at different wavelength bands, each spanning a limited spectral region (∼8.7 nm). (C) Combined emission signals across the lambda stack in each z-plane are linearly unmixed using reference emission spectral profiles from each Opal dye and lipofuscin to separate the contribution of individual fluorescent gene probes. Unmixed data is then projected across the z-axis. Single transcripts for SNAP25 (C), SLC17A7 (C’), GAD1 (C’’) and MBP (C''') (canonical markers for neurons, excitatory neurons, inhibitory neurons, and oligodendrocytes, respectively) can be separated from each other and from lipofuscin autofluorescence (C’'''). (D) Segmentation of unmixed fluorescent signals using dotdotdot. (E) Nuclear segmentation overlaid with transcript segmentation. (F) Masking with lipofuscin signal removes pixels confounded by autofluorescence from analysis. Green arrows highlight MBP+/SNAP25-/SLC17A7-/GAD1- oligodendrocytes. Red arrows highlight MBP-/SNAP25+/SLC17A7+/GAD1- excitatory neurons. Pink arrows highlight MBP-/SNAP25+/SLC17A7-/GAD1+ inhibitory neurons. Cyan double arrows highlight lipofuscin. Scale bar is 20um.