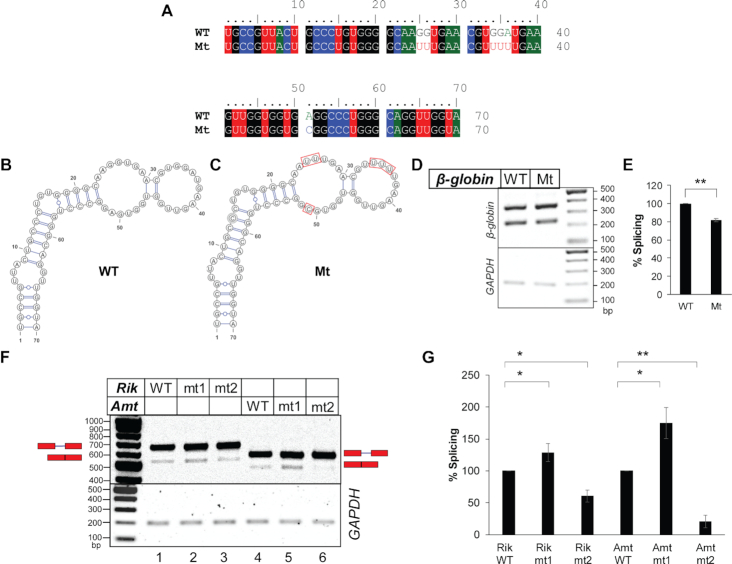
Figure 3.
Sequence- and context-specific effects of single-stranded nucleotide sequence in the exonic segments immediately upstream of the introns on splicing. (A) Sequence alignment of the 5′ exonic segment of WT and mutant β-globin variants. (B, C) Secondary structure models of the above exonic segment of WT β-globin (B) and the mutant β-globin (C) (both predicted by ‘RNAstructure’), which are identical to experimentally derived secondary structure of WT β-globin shown in Supplementary Figure S2A. (D) Transfection-based splicing assay of the WT and the mutant β-globin variants. (E) Plots of quantified band-intensities shown in (D) (n = 3); statistical significance was tested to validate if the splicing competence of Mt mutant of β-globin is less than that of WT β-globin; ‘*’0.005 < P≤ 0.05, ‘**’P≤ 0.005, N.S. = not significant; background-subtracted gel-images of two additional biological replicates are shown in Supplementary File 8. (F) Splicing products of mouse pre-mRNAs 2610507B11Rik IVS17 (Rik) and Amt IVS1 (Amt) and their mt1 and mt2 mutants (see Supplementary Figure S5) visualized in agarose gel (representative); positions of pre-mRNAs and mRNAs are marked and GAPDH amplification used as an internal control shown; the original ‘Image Lab’ image File is provided as Supplementary File 3. (G) A bar-graph showing comparison of spliced mRNAs normalized with total RNA generated from the transfected plasmids; error bars (n = 3) indicate standard deviation; statistical significance was tested to validate if mt1 mutants have higher and mt2 mutants have lower splicing competence than the WT substrate; ‘*’0.005 < P≤ 0.05, ‘**’P≤ 0.005; the background subtracted gel-images of two additional biological replicates used for densitometric analysis are shown in Supplementary File 8.