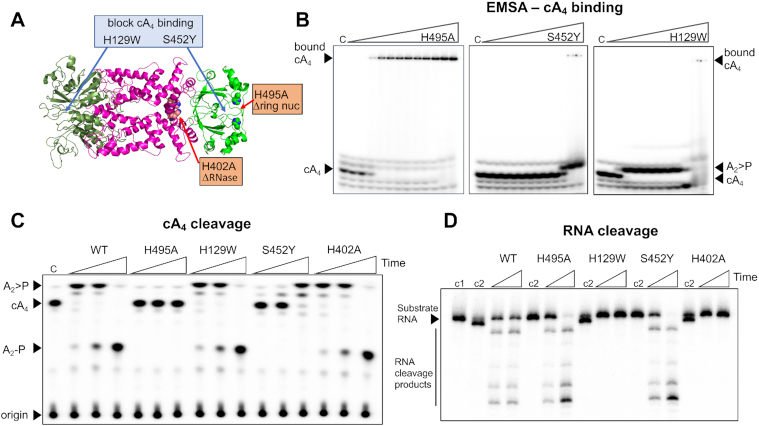
Figure 5.
Dissection of cA4 binding and catalysis by Csx1–Crn2. (A) Cartoon representation of Csx1–Crn2 showing sites targeted by mutagenesis. (B). Electrophoretic mobility shift assay (EMSA) showing binding of radiolabelled cA4 (∼10 nM) by Csx1-Crn2 variants (0.001, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.08, 0.1, 1.0, 10.0 and 20.0 μM, respectively). On each gel is a control reaction (c) with cA4 in the absence of protein. The ring nuclease defective variant H495A bound cA4 with an apparent dissociation constant around 20 nM. The S452Y variant, which was designed to abolish cA4 binding by the ring nuclease domain, had a severe binding defect, with cA4 degradation apparent at high protein concentrations. The H129W variant, designed to abolish cA4 binding in the CARF domain, showed cA4 degradation starting from around 20 nM enzyme. The data are representative of three technical replicates. (C) TLC showing degradation of cA4 (200 nM) by wild-type and variant enzymes (5 μM dimer) at 50°C. A time-course of 10 s, 1 min and 10 min was carried out and a control reaction (c) incubating buffer with radiolabeled cA4 for 10 min is also shown. The H495A variant was inactive as shown previously. The H129W and H402A variants targeting the CARF and HEPN (Csx1) domains, respectively, had normal ring nuclease activity. Whereas the S452Y variant targeting the Crn2 domain had strongly reduced activity, consistent with weaker binding of cA4 by the Crn2 domain. (D) Denaturing PAGE showing cleavage of radiolabelled substrate RNA (100 nM) by Csx1-Crn2 and variants (5 μM dimer) and 100 μM cA4 at 50°C. RNA cleavage was examined at two time-points (5 & 30 min) and control reactions incubating radiolabelled RNA in buffer (c1) or RNA and protein in the absence of cA4 activator (c2) for 30 min are also shown. The H129W and H402A variants were inactive as expected. The data are representative of three technical replicates.