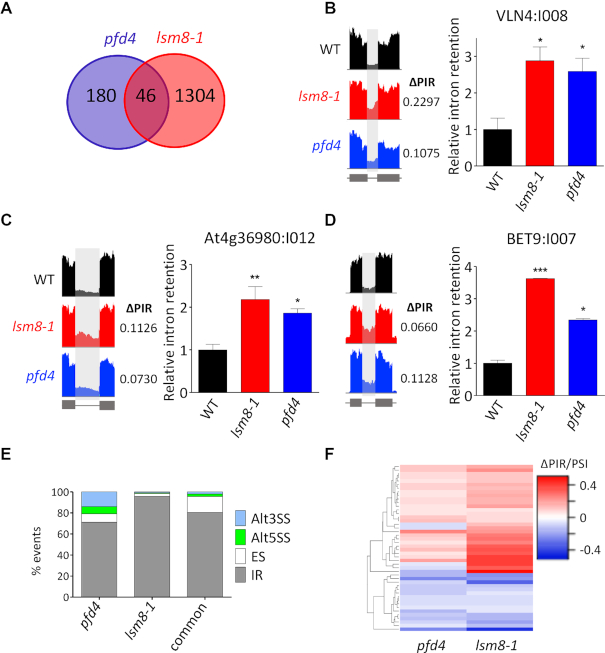
Figure 6.
Analysis of pre-mRNA splicing by RNA-seq in pfd4 and lsm8-1 mutants in response to cold treatment. (A) Venn diagram showing the number of splicing events altered in the pfd4 and lsm8-1 mutants. (B–D) Examples of common IR highlighted in IGV plots (left) and validated by RT-qPCR (right); VLN4:I008 (B), At4g36980:I012 (C), and BET9:I007 (D). (E) Percentage of the different splicing events affected in each mutant and in both. Alt3SS and Alt5SS, alternative 3′ and 5′ splice site, respectively; ES, exon skipping; IR, intron retention. (F) Hierarchical clustering of common events by ΔPIR/PSI (percent intron retention/percent spliced-in) value.