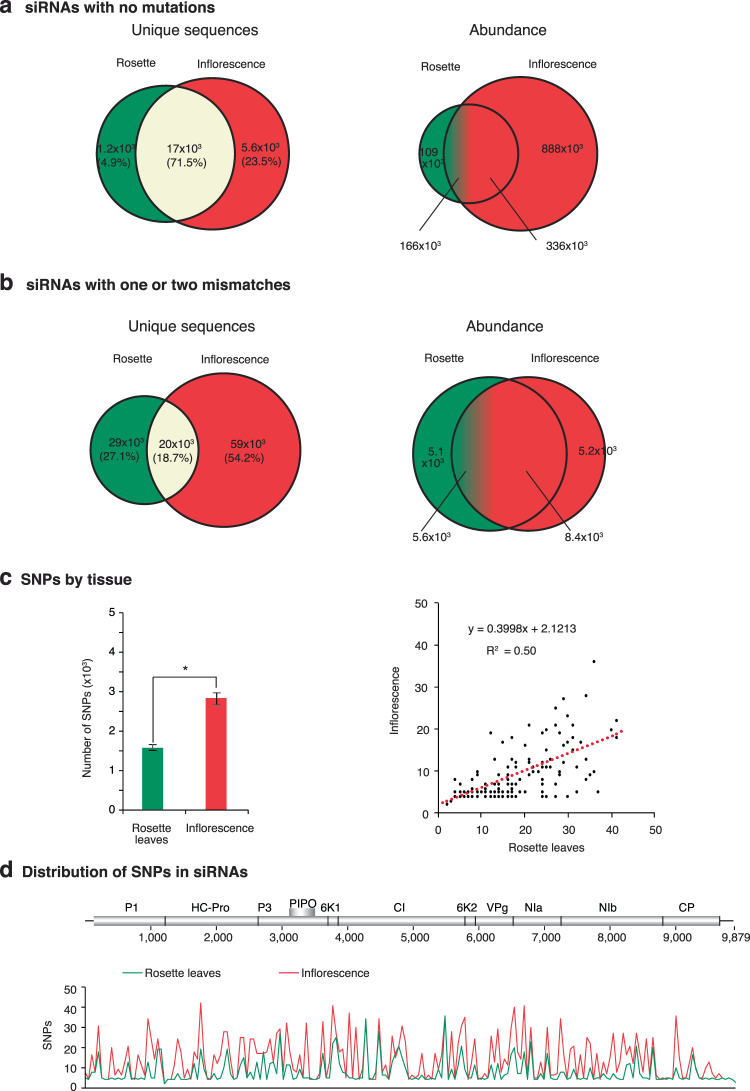
Figure 3.
Comparison of TuMV-derived siRNAs in inoculated rosette leaves and systemically infected inflorescence. 21- to 24-nt siRNAs were pooled and correspond to samples in Fig. 1. Unique sequences were counted per tissue and their abundance normalized to reads per million. Values are the average of two biological replicates. Virus-derived siRNAs with (a) no mutations and (b) with one or two mismatches. (c) Accumulation of single nucleotide polymorphisms (SNPs) in virus-derived siRNAs in inoculated rosette leaves, in systemically infected inflorescence, and their correlation. The asterisk (*) indicates significant difference (p-value < 0.001). (d) Genome-wide distribution of single nucleotide polymorphisms in virus-derived siRNAs estimated in a 50-nt window.