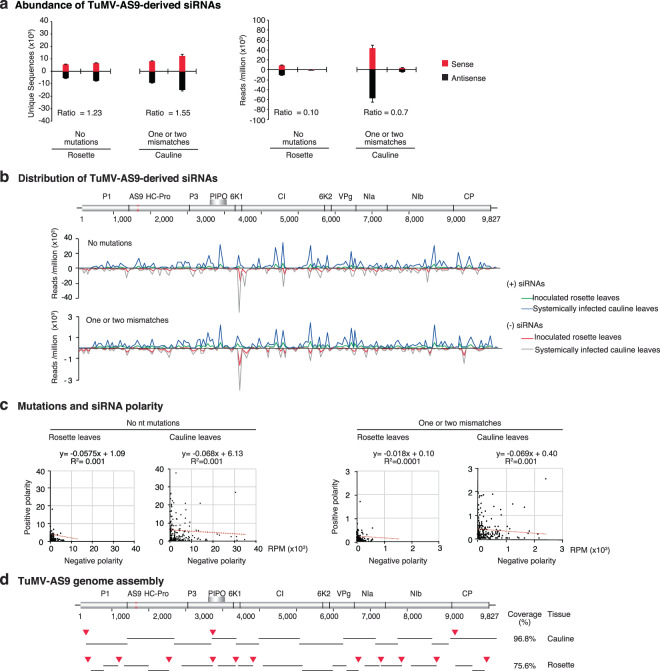
Figure 5.
Genome-wide distribution of siRNAs derived from suppressor-deficient TuMV-AS9 and de-novo genome assembly. siRNAs 21- to 24-nt were pooled and classified based on the number of mismatches. Numbers represent average and standard error from two biological replicates from inoculated rosette leaves (7 dpi) and systemically infected cauline leaves (15 dpi) of ago2-1 single mutant Arabidopsis. Number of reads were normalized to reads per million. (a) Unique sequences and their abundance by polarity. The ratio of siRNAs with mismatches to siRNAs with no mutations is indicated for each tissue. (b) Genome-wide distribution of virus-derived siRNAs by match class, polarity, and tissue. (c) Correlation between siRNAs of positive and negative polarity, by match class and tissue. (d) De-novo assembly of the TuMV-AS9 genome using siRNAs with no mutations. Coverage is expressed as percent of the genome. Red arrow heads point to gaps in the assembly.