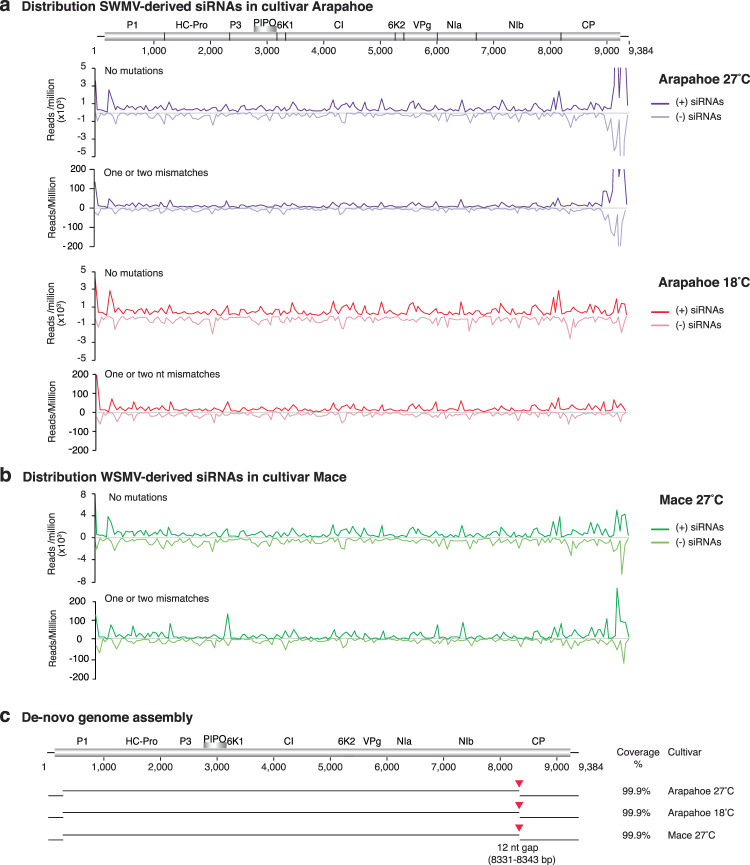
Figure 7.
Genome-wide distribution of Wheat streak mosaic virus (WSMV)-derived siRNAs and de-novo genome assembly. SiRNAs (21- to 24-nt) were pooled and correspond to two contrasting cultivars, Arapahoe (susceptible) at 18 °C and 27 °C and Mace (susceptible at high temperature) at 27 °C. Abundance was normalized to reads per million. (a) Genome-wide distribution of WSMV-derived siRNAs in cultivar Arapahoe, and (b) cultivar Mace. (c) De-novo assembly of the WSMV genome using siRNAs with no mutations. Genome coverage is indicated in percent. Red arrow heads point to gaps (12 nt) in assembly, which map to the coat protein from nt 8331 to 8343.