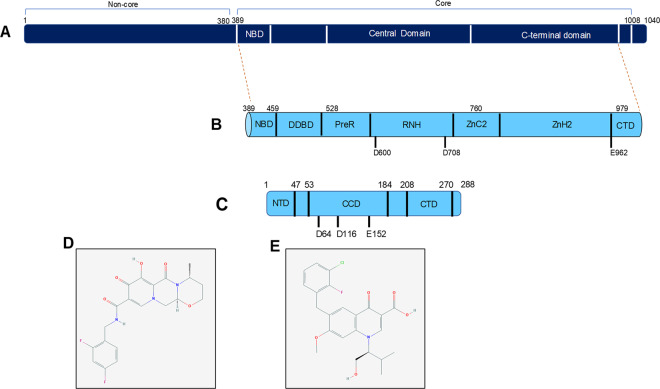
Fig. 1. Domain structure of RAG1 and HIV integrase and chemical structure of integrase inhibitors.
a, b Human RAG1 is made of 1040 amino acids. Core RAG1 (amino acids 389–1008) is used for in vitro experiments. Core RAG1 can be divided into three broad domains, namely nonamer binding domain (NBD; amino acids 389–459), central domain (amino acids 528–760) and C-terminal domain (amino acids 760–979). The catalytically active motif of RAG1 lies in the core region, with the D600 and D706 in the central domain and E962 in the C-terminal domain46,47. Using cryo-EM microscopy the core region of RAG1 is further characterised into various modules containing NBD (nonamer binding domain), DDBD (DNA binding and dimerisation domain), PreR, RNH, ZnC2, and ZnH2 followed by CTD46,47. c Human immunodeficiency virus integrase is a small protein consisting of 288 amino acids. It has an N-terminal domain (NTD; amino acids 1–47), catalytic central domain (CCD) (amino acids 53–184) and C-terminal domain (CTD; amino acids 208–270). The catalytically active motif is present in the central domain of HIV integrase and shares similarity with RAG148,49. d, e Chemical structure of Dolutegravir (d)50 and Elvitegravir (e)51 used in the study.