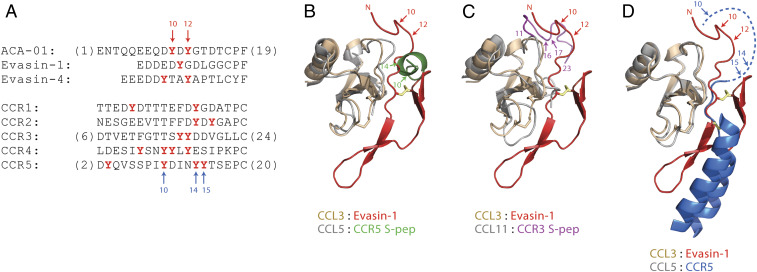
Fig. 7.
Sequence and structural comparisons of tyrosine sulfation sites in evasins and chemokine receptors. (A) Aligned partial sequences of the N-terminal regions of selected evasins (Top) and human CC chemokine receptors (Bottom). Alignment is based on the structural overlay of chemokine ligands in structures of evasin-1 bound to CCL3 and CCR5 bound to 5P7-CCL5 (D). Potentially sulfated tyrosine residues are in red bold type. (B–D) Partial structure of evasin-1 (residues 1–39 shown as red ribbons, with the first disulfide in yellow) bound to CCL3 (wheat ribbons, disulfides as sticks) (25) overlayed with (B) the structure of a CCR5-derived doubly sulfated peptide (CCR5 S-pep, green) bound to CCL5 (gray) (48), (C) the structure of a CCR3-derived doubly sulfated peptide (CCR3 S-pep, magenta) bound to CCL11 (gray) (47), and (D) the structure of CCR5 (transmembrane helices TM1 and TM7 shown as blue ribbons, with the connecting disulfide in yellow) bound to 5P7-CCL5 (gray) (25, 46). The dashed blue line indicates the N-terminal region of CCR5, which is not defined in the crystal structure. Numbered arrows indicate the known or likely positions of Tyr sulfation. The structural overlays are based on chemokine sequence alignments extending from the CC motif to end of the C-terminal α-helix. Disordered regions of chemokines and peptides are omitted for clarity.