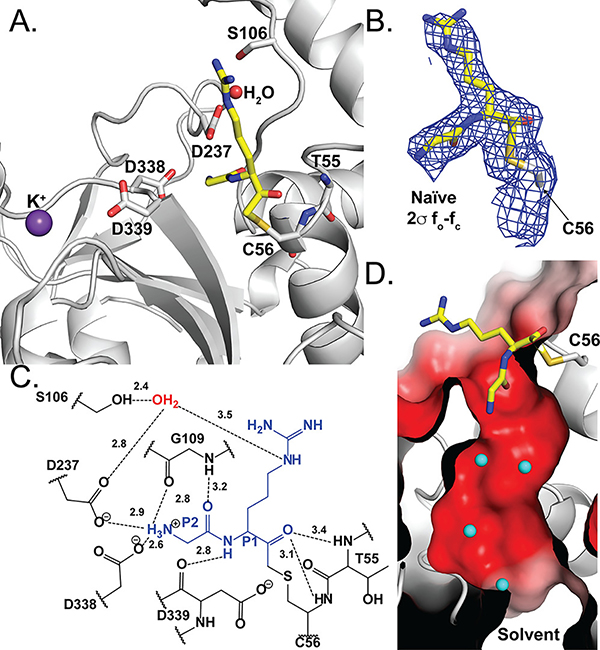
Fig. 3.
Pd_dinase in complex with the dipeptide inhibitor GR-AOMK determined by co-crystallization. (A) Cartoon representation of the residues and ordered water (red sphere) within the Pd_dinase:GR-AOMK structure. GR-AOMK (yellow carbon), covalently bound to the active Cys56, and active site residues (grey carbon) are shown as sticks (red oxygen, blue nitrogen, mustard sulfur). (B) The naïve fo-fc density map (blue) contoured at 2σ clearly indicated the orientation of the GR-AOMK dipeptide and covalent attachment to Cys56. (C) The hydrogen bonding network between GR-AOMK (blue), ordered water (red), and Pd_dinase active site residues (black) show that the majority of interactions with the inhibitor are to the N-terminal P2 Gly residue. (D) The surface electrostatic potential of the Pd_dinase active site consists of a highly electronegative channel that connects the solvent-exposed surface with the negatively charged active site, as shown with a cross-sectional representation. Ordered water molecules (cyan) are found throughout the channel. The electrostatic potential is colored: blue, positive potential (10 mV); white, neutral potential (0 mV); and red, negative potential (−10 mV).