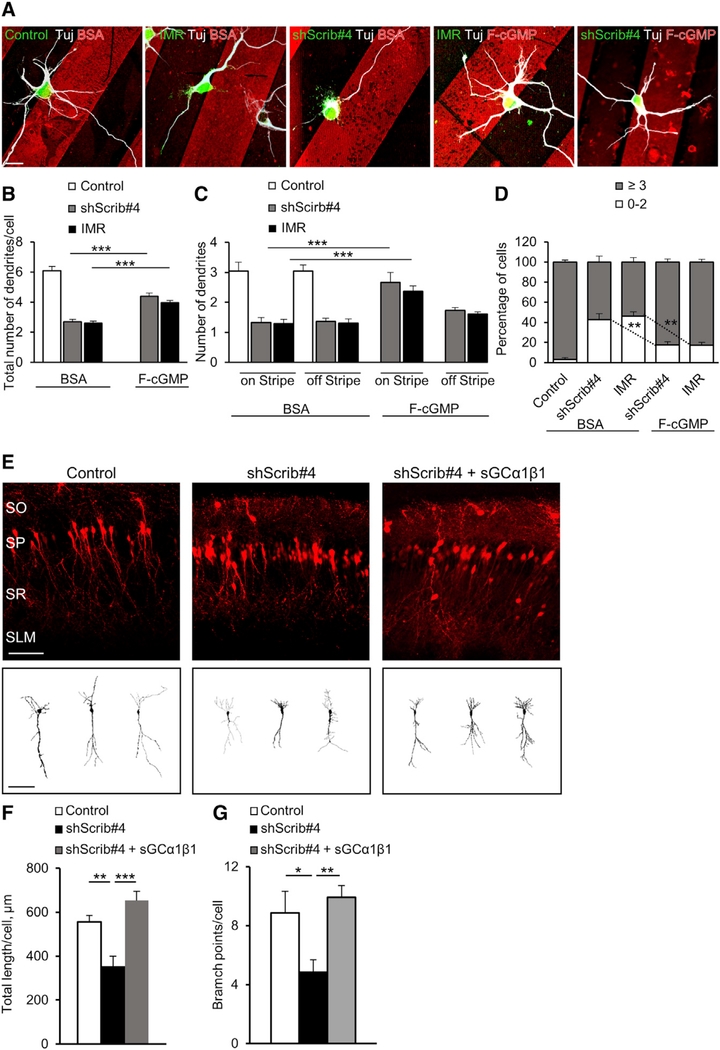
Figure 6. Local cGMP Elevation or sGC Expression Rescued Dendrite Defects following Scribble Knockdown.
(A) Representative images of hippocampal neurons, transfected with ShScrib#4 or control-shRNA, or EGFP-fused IMR or control vector, plated on substrates coated with stripes of membranepermeable fluorescent analog of cGMP (F-cGMP, red) or control fluorescently conjugated BSA (5 DIV) immunostained with Tuj-1. Shown are neurons with their soma located at the stripe boundary. Scale bar represents 20 μm.
(B and C) Quantification of average total dendrite number per cell (B) or dendrite number initiated in the “on-stripe” or “off-stripe” region of F-cGMP or BSA stripe (C), for polarized neurons with their soma located at the stripe boundary (5 DIV) for cells transfected with ShScrib#4 or control-shRNA, or IMR or control vector (n = 3–5 cultures, 50–75 cells each; one-way ANOVA, Tukey’s multiple comparison test; ***p ≤ 0.001).
(D) Quantification of the average percentage of cells with 0–2 or more than 3 dendrites (5 DIV) for the same dataset as in (B) and (C) (one-way ANOVA, Tukey’s multiple comparison test, **p ≤ 0.01).
(E) Representative images of P7 rat CA1 pyramidal neurons, transfected in utero with control-shRNA, or ShScrib#4 alone or together with functional sGC enzyme (rat sGC-α1 and -β1 subunits), to test for rescue of Scribble knockdown with sGC. Scale bar represents 100 mm. Bottom, sample tracings of neuritic arbor of representative neurons. Scale bar represents 20 μm.
(F and G) Quantification of average total apical dendrite length (F) or branch points (G) per cell, for CA1 pyramidal neurons transfected with control-shRNA or ShScrib#4 alone or with reconstructed sGC (n = 20–30 cells; one-way ANOVA, Tukey’s multiple comparison test, *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001).
*p < 0.05, **p ≤ 0.01, ***p ≤ 0.001. Error bars represent SEM.