Figure 5. Polymer models aid in understanding the interactions that drive nuclear organization.
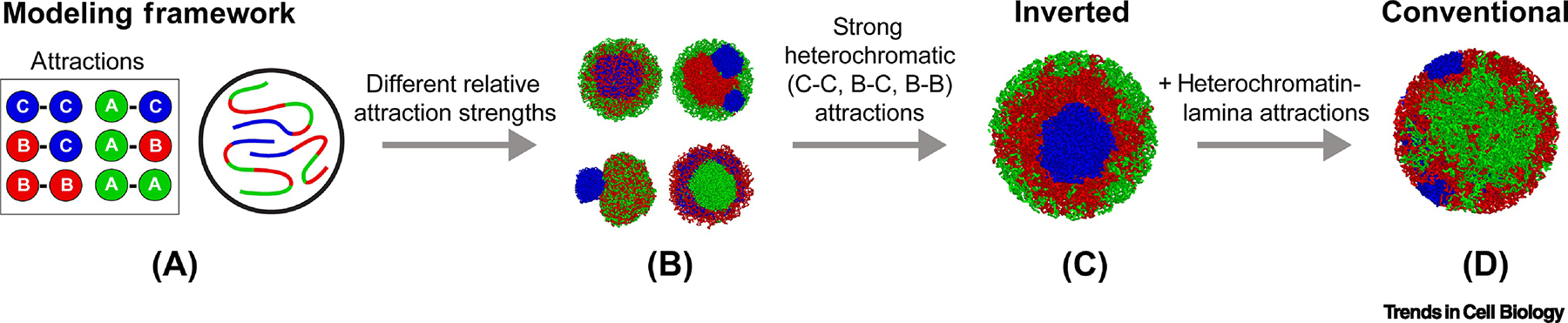
Models of chromosomes are built with confined polymers of three monomer types: A, B and C, representing EC, HC and cHC, respectively. A pair of monomers attract each other with different strengths depending on the monomer types involved (far left). Different relative strengths of the attractions result in a wide array of possible chromatin organizations (mid-left). Only certain relative strengths of attraction between monomers result in an inverted arrangement (mid-right). The same simulation parameters, but complemented with attraction of HC and cHC to the nuclear lamina, successfully recapitulate the conventional arrangement (far right).
