Figure 1. Traditional Targets.
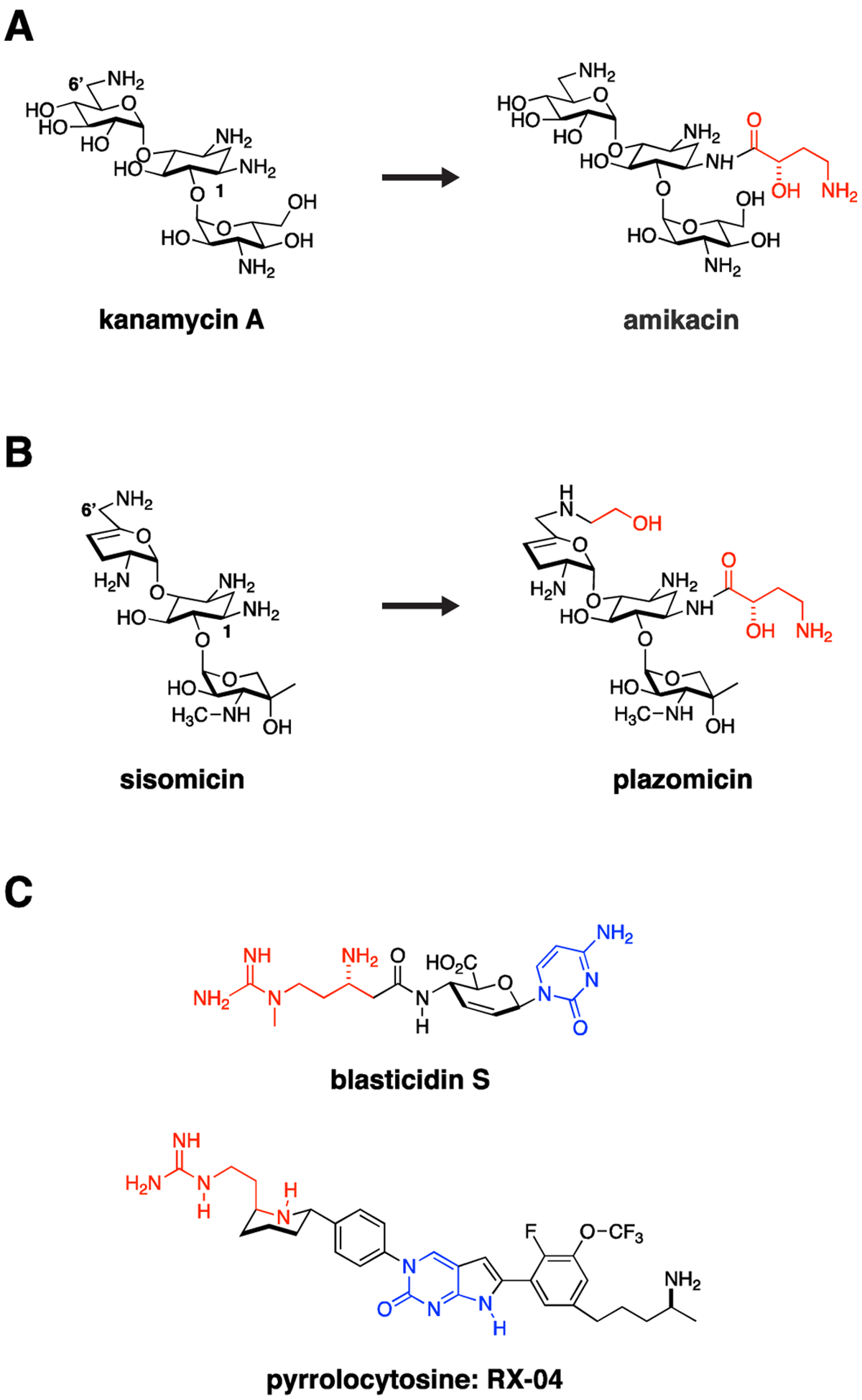
The ribosome and bacterial cell wall remain tried and true targets for Gram-negative antimicrobials. Resistance often occurs through acquisition of antibiotic modifying enzymes or hydrolases. Medicinal chemistry strategies can be used to overcome these liabilities by altering antibiotics to impede action of these resistance enzymes. For example, (A) kanamycin was modified through addition of a hydroxyaminobutyric (HABA) acid (colored red) to the deoxystreptamine ring to yield amikacin. The HABA group sterically blocks several aminoglycoside-modifying enzymes, significantly broadening the activity spectrum of amikacin, which in doing so may preserve activity against CRE strains. (B) This strategy was taken one step further in development of plazomicin from sisomicin with both hydroxyethyl and HABA modifications to the parent sisomicin shown in red. This constellation of changes makes plazomicin immune to essentially all aminoglycoside modifying enzymes circulating in CRE. Unfortunately, plazomicin is not able to overcome emerging 16S ribosomal RNA methylasetransferase-based aminoglycoside resistance. (C) The pyrrolocytosine series is an example of a new class of antimicrobials modeled in part on the interaction of blasticidin S with the ribosome but extending into unique ligand binding space. These molecules share a 1,3-aminoguanidine (red) and a cytosine ring system (blue). The presumption from this strategy is that pre-existing resistance enzymes that modify either drug or the binding target in the ribosome should not have previously evolved, thereby extending the useful lifetime of the antimicrobial.
