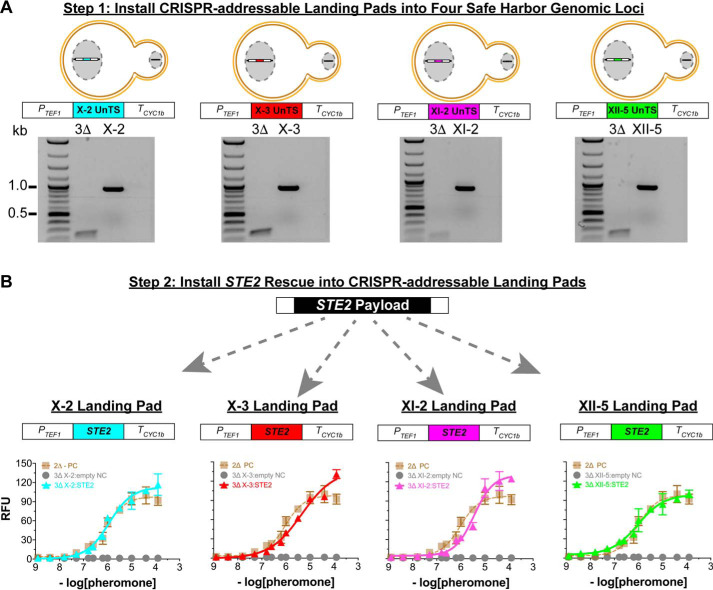
Figure 2.
Installation and validation of individual CRISPR-addressable landing pads. A, top panel, The variants of our 3Δ reporter strain, each containing a single landing pad placed at known safe harbor chromosome loci X-2, X-3, XI-2, or XII-5. Each landing pad contains a 20-bp UnTS not found in the native yeast genome that provides a synthetic locus for CRISPR-addressable editing. A, bottom panel, PCR validation of landing pad installation. PCR primers used were homologous to the native genomic loci sequences flanking the landing pads. The expected product sizes were 915 bp for X-2, X-3, and XII-5 landing pads and 816 bp for the XI-2 landing pad. B, rescuing pheromone signaling by expressing Ste2 from each landing pad in the single-padded 3Δ reporter strains. The data are reported as relative fluorescence units (RFU) at an A600 nm of 1.0 and instrument gain of 1200. Each strain was titrated with α-factor, the endogenous peptide pheromone for Ste2. Triangles correspond to strains expressing Ste2 from a landing pad: X-2 (turquoise), X-3 (red), XI-2 (purple), and XII-5 (green). Positive control titrations (labeled PC) correspond to the 2Δ reporter strain with Ste2 expressed from its native genome locus. Negative control titrations (labeled NC) correspond to the empty-padded 3Δ reporter strains described for A. Error bars represent the S.D. of four biological replicates.