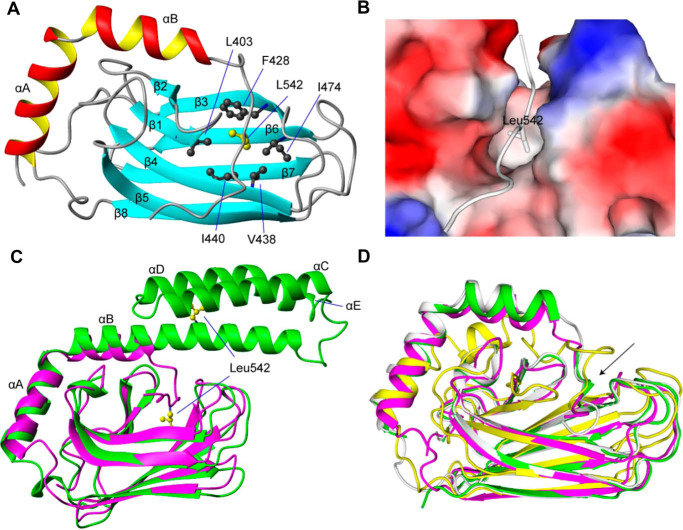
Figure 5.
The NMR structure of glutathionylated hHsp70 SBD. A, ribbon representation of the glutathionylated hHsp70 SBD. Residues 611–641, which are disordered in the unmodified native structure, are not displayed. B, molecular surface of the glutathionylated hHsp70 SBD and binding of residue Leu-542 of the unraveled α-helix B within the hydrophobic core. C, superimposition of glutathionylated hHsp70 SBD (residues 385–641; purple) and nonglutathionylated hHsp70 SBD (residues 385–616, PDB code 4PO2; green). D, superimposition of glutathionylated hHsp70 SBD (residues 385–641; magenta) and truncated Hsp70 SBD domains of different species: rat Hsp70 (residues 383–542, PDB code 1CKR; yellow), human Hsp70 (residues 393–543, PDB code 4WV5; green), and bovine Hsp70 (residues 1–554, PDB code 1YUW; white). The arrow indicates the segments of the SBDα that have collapsed into the SBDβ.