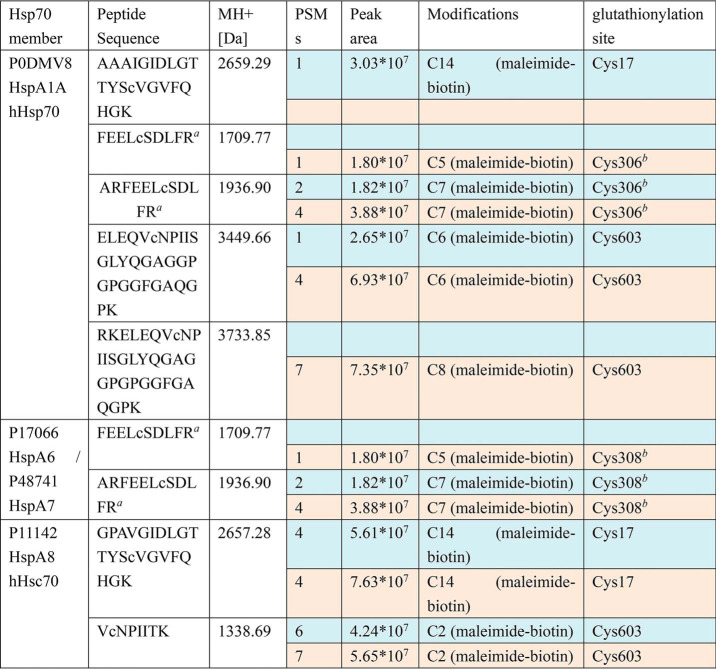
Table 1.
Representative results from one round of HeLa cell preparation followed by biotin switch enrichment and nanoLC-LTQ-Orbitrap XL MS/MS detection, showing identified glutathionylation sites on different Hsp70 family members in untreated (blue rows) and diamide-treated cells (beige rows)
a Source data have been deposited into the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD017717. Spreadsheet S1 contains MS data sets for human Hsp70 (HspA) glutathionylation as follows: Data set 1, sample 1 untreated (blue rows in Table 1); Data set 2, sample 1 treated with 10 mm diamide (beige rows in Table 1); Data set 3, sample 2 untreated (MS detection 1 of 2); Data set 4, sample 2 untreated (MS detection 2 of 2); Data set 5, sample 2 treated with 1 mm diamide (MS detection 1 of 2); Data set 6, sample 2 treated with 1 mm diamide (MS detection 2 of 2); Data set 7, sample 3 untreated (MS detection 1 of 2); Data set 8, sample 3 untreated (MS detection 2 of 2).
b Peptide is found in both hHsp70 (HspA1A) and in HspA6/A7, so it cannot be unambiguously assigned.