Figure 7.
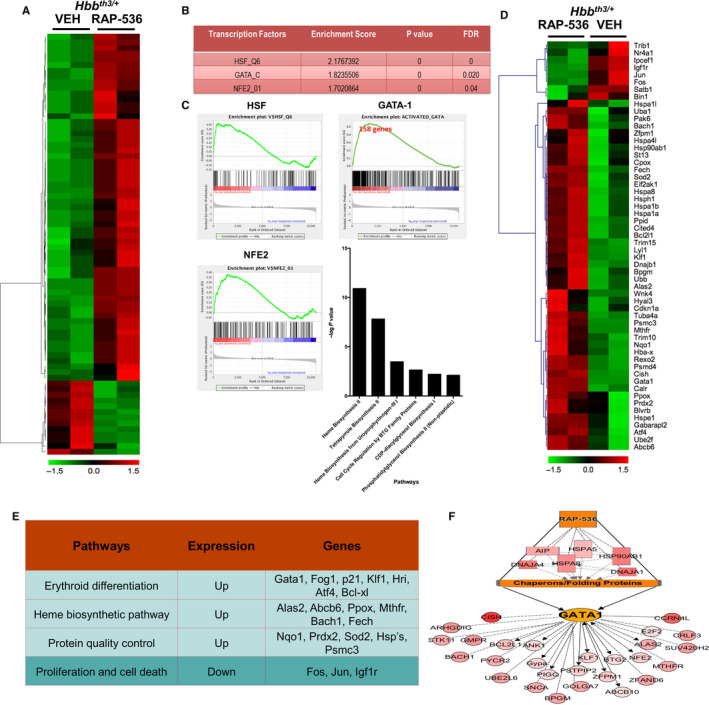
RAP‐536 up‐regulates Gata1 and its target gene signature in erythroid precursors of Hbbth3/+ β‐thalassaemic mice. A, Heat map for RNA sequencing analysis of sorted erythroid precursors from Hbbth3/+ mice depicting differentially regulated genes 16 h after treatment with a single dose of RAP‐536 (30 mg/kg, i.p) or vehicle. B, Main transcription factors up‐regulated in erythroid precursors by RAP‐536 treatment. FDR—false discovery rate. C, Gene set enrichment analysis to identify those differentially regulated in erythroid precursors by RAP‐536 treatment, showing plots for GATA‐1, HSF and NFE2. Bar graph indicates pathways that were significantly activated by GATA‐1 in erythroid precursors by RAP‐536 treatment. D, Representative heat map with each column representing genes up‐regulated (red) or down‐regulated (green) in erythroid precursors of an individual mouse by treatment with RAP‐536 or vehicle (VEH). E, Pathways affected by RAP‐536 treatment and some key genes involved. F, Linkage analysis model depicting RAP‐536‐induced changes in gene expression (all up‐regulated) in erythroid precursors
