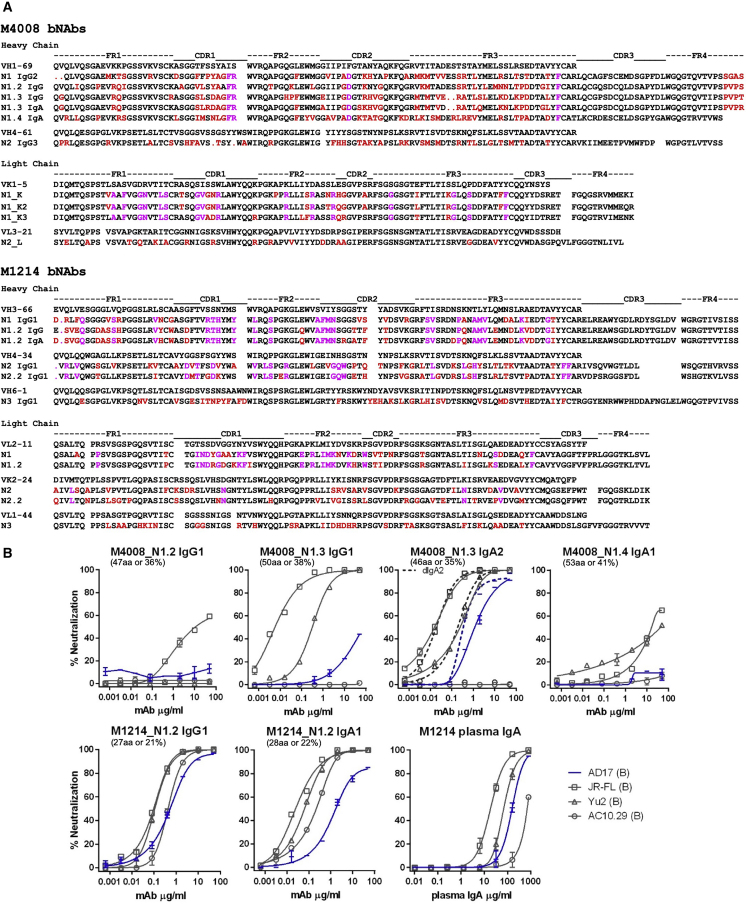
Figure 3.
Sequence Alignment and Neutralization Assessment of Representative bNAb Lineage Members
(A) The deduced aa sequences of the variable region of bNAbs and representative lineage members are shown. Framework (FR) and complementarity-determining regions (CDRs) are indicated above the sequence alignment. The top sequence in each group represents the deduced germline V-gene sequence. The dot symbol marks deletion; red residues indicate somatic hypermutations, and magenta residues indicate common somatic hypermutations among lineage members, as shown.
(B) The bNAb lineage members were reconstituted as IgG1, IgA1, IgA2, or dIgA2, and then assessed for neutralization against the indicated Env pseudoviruses. The number and percent of aa difference from the probe-identified bNAb reference is indicated in parenthesis for the heavy-chain variable region of each clonal member. The M4008_N1 clonal members shown were paired with N1_K2 κ chain, and the M1214_N1.2 clonal members shown were paired with N1.2 λ chain. Also shown is the neutralization profile of the purified IgA fraction (polyclonal) from donor M1214 plasma. The VSVAD17 probe strain AD17 is highlighted in blue. Neutralization was performed in duplicate wells, and the data shown are means with SEM.