Figure 5.
Cryo-EM Structure of M1214_N1 Fab in Complex with CH505 SOSIP Trimer
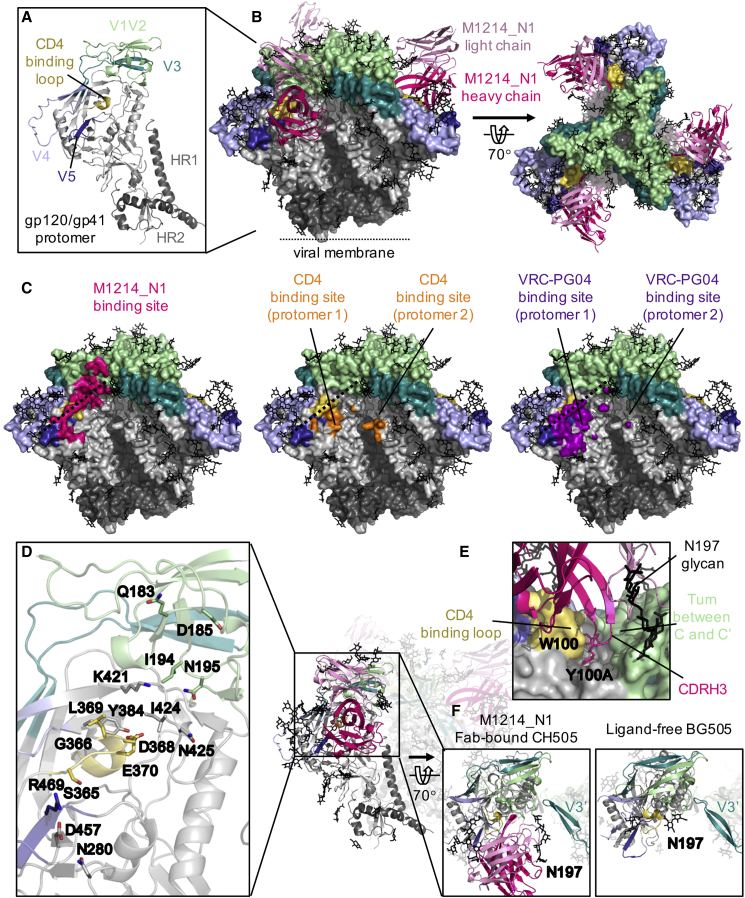
(A) Structural model of a CH505 gp120/gp41 protomer. The variable (V1-V5) regions and CD4-binding loop in gp120 and HR1 and HR2 helices in gp41 are highlighted, with the same color scheme throughout the figure.
(B) Cryo-EM structure of three M1214_N1 Fabs (ribbon) on a CH505 SOSIP trimer in side (left) and top (right) views. Glycans on the trimer are depicted in black sticks. For clarity, only the Fv domains of M1214_N1 are shown.
(C) Comparison of the binding site of M1214_N1 (hot pink) to that of CD4 (orange) (Protein Data Bank [PDB]: 5U1F) and the CD4bs-directed bNAb VRC-PG04 (purple) (PDB: 3J5M). The dashed line depicts the CD4-binding loop ridge.
(D) Env residues with surface contact areas > 10 Å2 in M1214_N1 binding are shown in sticks.
(E) Close-up view of the pocket accommodating CDR H3 of M1214_N1.
(F) Position change of the N197 glycan on M1214_N1-bound CH505 SOSIP compared with a ligand-free BG505 SOSIP (PDB: 4ZMJ), restricting the movement of the V3 loop from the neighboring protomer (V3′).