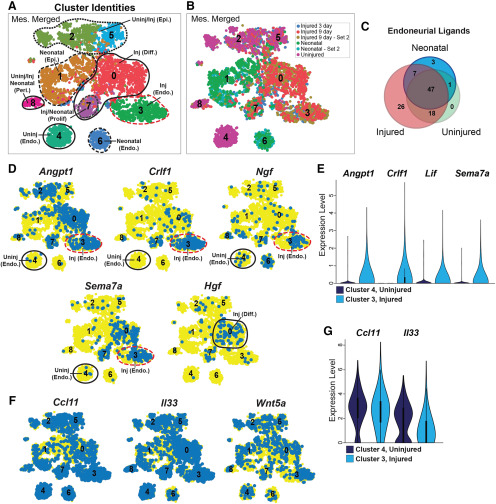
Figure 4.
Characterization of ligand expression in sciatic nerve mesenchymal cells (see also Extended Data Fig. 4-1). Pdgfra-positive mesenchymal cell transcriptomes from the injured, uninjured and neonatal nerve datasets (in Figs. 1E, 2C,E; also see Carr et al., 2019) were extracted, combined together, analyzed and batch-corrected using Harmony data integration and clustered in Seurat based on principal components. A, B, t-SNE visualizations of the combined mesenchymal cells showing clusters (A) and datasets of origin (B). The clusters in A are also annotated based on marker gene expression (Extended Data Fig. 4-1) and datasets of origin. Neonatal and 9 d refer to the FAC-sorted preparations at these timepoints, while neonatal – set 2 and 9 d – set 2 refer to the cells prepared with myelin removal beads. C, Venn diagram showing the overlap of ligand mRNAs expressed in neonatal (cluster 6), injured (cluster 3), and uninjured (cluster 4) nerve endoneurial mesenchymal cells from the combined mesenchymal nerve dataset in A. Ligand mRNAs were considered to be expressed if they were detectable in 2% or more cells in the relevant cluster. D, t-SNE gene expression overlays of the combined mesenchymal cell data in A for Angpt1, Crlf1, Ngf, Sema7a, and Hgf. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. Relevant clusters are circled and annotated, including uninjured endoneurial (Uninj Endo.), injured endoneurial (Inj Endo.), and injured differentiating (Inj Diff.) mesenchymal cells. E, Violin plots showing the relative expression of Angpt1, Crlf1, Lif, and Sema7a in injured endoneurial mesenchymal cell cluster 3 versus uninjured endoneurial cell cluster 4 from the dataset in A. F, t-SNE gene expression overlays of the combined mesenchymal cell data in A for Ccl11, Il33, and Wnt5a. Cells that detectably express the ligand are colored blue, and the numbers correspond to the clusters. G, Violin plots showing the relative expression of Ccl11 and Il33 in injured endoneurial mesenchymal cell cluster 3 versus uninjured endoneurial cell cluster 4 from the dataset in A.