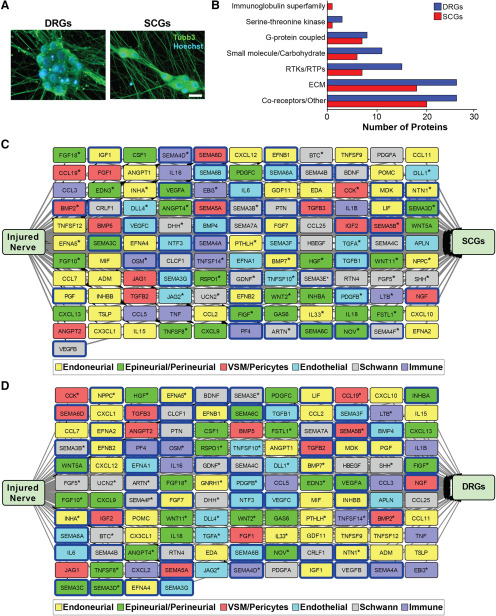
Figure 5.
Sensory and sympathetic neuron receptor expression and modeling of their predicted interactions with injured nerve-derived ligands (see also Extended Data Fig. 5-1). A, Images of E15 rat DRG sensory neurons cultured for 9 d (left panel) and neonatal rat SCG sympathetic neurons cultured for 6 d (right panel), immunostained for βIII-tubulin (Tubb3; green). Cells were counterstained with Hoechst 33258 to show cell nuclei (blue). Scale bars = 20 μm. B, Bar graphs showing types of receptor proteins detected on the surface of sensory and sympathetic neurons following cell-surface capture mass spectrometry. Classification of receptors was based on GO terms identified via the UniProt database (http://uniprot.org) as well as manual curation of all detected cell-surface proteins. RTKs/RTPs = receptor tyrosine kinases and receptor tyrosine phosphatases; ECM = extracellular matrix. C, D, Models showing predicted unidirectional paracrine interaction networks between the 143 injured nerve ligands and receptors expressed by sympathetic (C, SCGs) and sensory (D, DRGs) neurons, as defined at the transcriptional and proteomics levels. Interactions were predicted by computational modeling using the ligand-receptor database, and then manually curated for well-validated interactions. Nodes represent ligands that are color coded to identify the injured nerve cell type with the highest expression of the ligand mRNA based on the scRNA-seq analysis. Predicted interactions where the receptors were detected at the protein level are indicated by a blue box around the corresponding ligand node. Asterisks indicate ligands that were expressed at 4-fold higher levels in the indicated cell type as compared with all other injured nerve cell types. Arrows indicate directionality of interactions.