Abstract
A new coronavirus SARS-CoV-2, also called novel coronavirus 2019 (2019-nCoV), started to circulate among humans around December 2019, and it is now widespread as a global pandemic. The disease caused by SARS-CoV-2 virus is called COVID-19, which is highly contagious and has an overall mortality rate of 6.35% as of May 26, 2020. There is no vaccine or antiviral available for SARS-CoV-2. In this study, we report our discovery of inhibitors targeting the SARS-CoV-2 main protease (Mpro). Using the FRET-based enzymatic assay, several inhibitors including boceprevir, GC-376, and calpain inhibitors II, and XII were identified to have potent activity with single-digit to submicromolar IC50 values in the enzymatic assay. The mechanism of action of the hits was further characterized using enzyme kinetic studies, thermal shift binding assays, and native mass spectrometry. Significantly, four compounds (boceprevir, GC-376, calpain inhibitors II and XII) inhibit SARS-CoV-2 viral replication in cell culture with EC50 values ranging from 0.49 to 3.37 µM. Notably, boceprevir, calpain inhibitors II and XII represent novel chemotypes that are distinct from known substrate-based peptidomimetic Mpro inhibitors. A complex crystal structure of SARS-CoV-2 Mpro with GC-376, determined at 2.15 Å resolution with three protomers per asymmetric unit, revealed two unique binding configurations, shedding light on the molecular interactions and protein conformational flexibility underlying substrate and inhibitor binding by Mpro. Overall, the compounds identified herein provide promising starting points for the further development of SARS-CoV-2 therapeutics.
Subject terms: Structural biology, Molecular biology
Introduction
An emerging respiratory disease COVID-19 started to circulate among human in December 2019. Since its first outbreak in China from an unknown origin, it quickly became a global pandemic. As of May 26, 2020, there are 343,562 deaths among 5,406,282 confirmed cases in 215 countries (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The etiological pathogen of COVID-19 is a new coronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also called novel coronavirus (nCoV-2019). As the name indicates, SARS-CoV-2 is similar to SARS-CoV, the virus that causes severe acute respiratory symptoms in human and killed 774 people among 8098 infected worldwide in 2003.1 SARS-CoV-2 shares ~82% of sequence identity with SARS-CoV and to a less extent with Middle East respiratory syndrome coronavirus (MERS-CoV) (~50%).2,3 SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus that belongs to the β-lineage of the coronavirus.4 The β-lineage also contains two other important human pathogens, the SARS-CoV and MERS-CoV. The mortality rate of COVID-19 is around 6.35% as of May 26, 2020, which is lower than that of SARS (~10%) and MERS (~34%).1 However, current data indicate that COVID-19 is more contagious and has a larger R0 value than SARS and MERS5, resulting in higher overall death tolls than SARS and MERS. The SARS-CoV-2 virus is currently spreading at an alarming speed in Europe and the United States.
There is currently no antiviral or vaccine for SARS-CoV-2. The SARS-CoV-2 viral genome encodes a number of structural proteins (e.g., capsid spike glycoprotein), non-structural proteins (e.g., 3-chymotrypsin-like protease (3CL or main protease), papain-like protease, helicase, and RNA-dependent RNA polymerase), and accessary proteins. Compounds that target any of these viral proteins might be potential antiviral drug candidates.6,7 In this study, we focus on the viral 3CL protease, also called the main protease (Mpro), and aim to develop potent Mpro inhibitors as SAR-CoV-2 antivirals. The Mpro plays an essential role in coronavirus replication by digesting the viral polyproteins at more than 11 sites, and it appears like a high-profile target for antiviral drug discovery.8–11 The Mpro has a unique substrate preference for glutamine at the P1 site (Leu-Gln↓(Ser, Ala, Gly)), a feature that is absent in closely related host proteases, suggesting that it is feasible to achieve high selectivity by targeting viral Mpro. As such, we developed the fluorescence resonance energy transfer (FRET)-based enzymatic assay for the SARS-CoV-2 Mpro and applied it to screen a focused library of protease inhibitors. Here we report our findings of several novel hits targeting SARS-CoV-2 Mpro and their mechanism of action. The in vitro antiviral activity of the hits was also evaluated in cell culture using infectious SARS-CoV-2 virus. Overall, our study provides a list of drug candidates for SARS-CoV-2 with a confirmed mechanism of action, and the results might help speed up the drug discovery efforts in combating COVID-19. The compounds identified herein represent some of the most potent and selective SARS-CoV-2 Mpro inhibitors so far with both enzymatic inhibition and cellular antiviral activity.8,10,11 The X-ray crystal structure of SARS-CoV-2 Mpro with GC-376 showed that the compound can adopt two configurations R and S, offering a molecular explanation of the high binding affinity of the aldehyde-containing inhibitors. Significantly, the discovery of calpain II and XII inhibitors as potent SARS-CoV-2 antivirals suggests that it might be feasible to design dual inhibitors against the viral Mpro and the host calpains/cathepsins, both of which are important for viral replication.
Results
Establishing the FRET-based assay for the SARS-CoV-2 Mpro
The Mpro gene from SARS-CoV-2 strain BetaCoV/Wuhan/WIV04/2019 was inserted into pET-29a(+) vector and expressed in BL21(DE3) Escherichia Coli. with a His-tag in its C-terminus. The Mpro protein was purified with Ni-NTA column to high purity (Fig. 1a). The determined molecular weight of Mpro is 34,861 Da from native mass spectrometry, which matches the calculated molecular weight of Mpro without the N-terminal methionine (Supplementary information, Fig. S1). The first methionine might be cleaved by the methionine aminopeptidase as reported before.12 To establish the FRET assay condition, we designed a FRET based substrate using the sequence between viral polypeptide NSP4-NSP5 junction from SARS-CoV-2: Dabcyl-KTSAVLQ/SGFRKME(Edans).8 We then tested the Mpro proteolytic activity in buffers with different pH. We found that Mpro displays the highest activity in pH 6.5 buffer (Fig. 1b), which contains 20 mM HEPES, 120 mM NaCl, 0.4 mM EDTA, 4 mM 1,4-dithiothreitol (DTT) and 20% glycerol. As such, all the following proteolytic assay was conducted using this pH 6.5 buffer. A standard curve was generated to convert the relative fluorescence unit (RFU) to the amount of the cleaved substrate (nM) such that kcat can be calculated (Fig. 1c). Next, we characterized the enzymatic activity of this SARS-CoV-2 Mpro by measuring the Km and Vmax values. When 200 nM Mpro was mixed with various concentration of FRET substrate (0–100 µM), the initial velocity was measured and plotted against substrate concentration. Curve fitting with Michaelis–Menten equation gave the best-fit values of Km and Vmax as 28.2 ± 3.4 µM and 31.7 ± 1.6 RFU/s, respectively (Fig. 1d). The calculated kcat/Km was 5,624 s−1 M−1, which is similar to the previously reported value of 3,426.1 s−1 M−1 by Hilgenfeld et al.8, suggesting that the C-terminal His-tag Mpro is as active as the native Mpro.
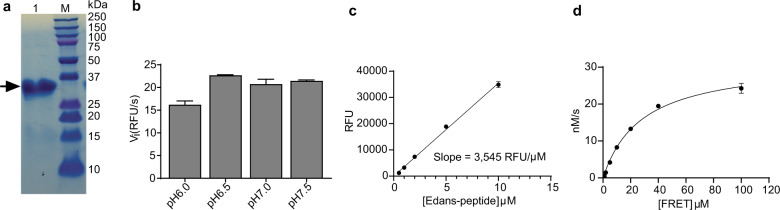
Fig. 1. SARS-CoV-2 Mpro expression and characterization.
a SDS-PAGE of His-tagged Mpro (lane 1); Lane M, protein ladder; the calculated molecular weight of the His-tagged Mpro is 34,861 Da. b Reaction buffer optimization: 250 nM His-tagged Mpro was diluted into three reaction buffers with different pH values. c An Edans standard curve was generated to convert RFU to the amount of the cleaved substrate (nM). d Michaelis–Menten plot of 200 nM His-tagged Mpro with various concentrations of FRET substrate in pH 6.5 reaction buffer. The best-fit Vmax = 31.7 ± 1.6 nM/s; Km = 28.2 ± 3.4 µM, and the calculated kcat/Km = 5,624 s−1 M−1.
Primary screening of a focused protease inhibitor library against the SARS-CoV-2 Mpro
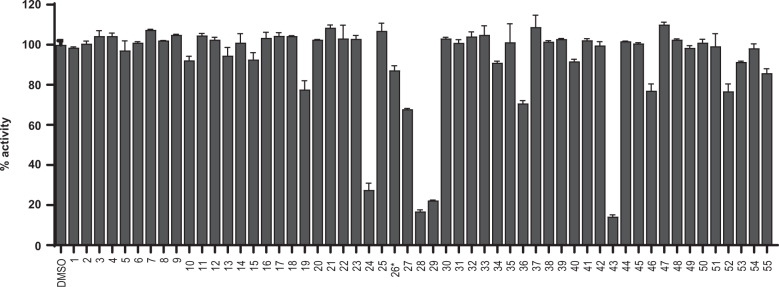
With the established FRET assay condition, we screened a collection of protease inhibitors from the Selleckchem bioactive compound library to identify potential SARS-CoV-2 Mpro inhibitors (Table 1). The protease inhibitors are grouped based on their targets and mechanism of action, including proteasome inhibitors (1–8), HIV protease inhibitors (9–14), γ-secretase inhibitors (15–22), HCV NS3–4A protease inhibitors (23–29), DPP-4 inhibitors (30–35), miscellaneous serine protease inhibitors (36–39), cathepsin and calpain protease inhibitors (40–43), miscellaneous cysteine protease inhibitors (44–48), matrix metalloprotease inhibitors (49–51), and miscellaneous protease inhibitors (52–55). The inhibitors were pre-incubated with 100 nM of Mpro at 30 °C for 30 min in the presence of 4 mM DTT before the addition of 10 µM FRET substrate. The addition of DTT was to quench non-specific thiol reactive compounds and to ensure that Mpro is in the reducing condition. All compounds were tested at 20 µM, except compound 26, which was tested at 2 µM due to its fluorescent background. Encouragingly, four inhibitors (24, 28, 29 and 43) showed more than 60% inhibition against Mpro at 20 µM (Fig. 2). Among the hits, simeprevir (24), boceprevir (28), and narlaprevir (29) are HCV NS3–4A serine protease inhibitors, and compound MG-132 (43) inhibits both proteasome and calpain.
Table 1.
List of protease inhibitors tested against SARS-CoV-2 Mpro in the primary FRET assay.
 | |
 |
Fig. 2. Screening of known protease inhibitors against SARS-CoV-2 Mpro using the FRET assay.
20 µM of compounds (26 was tested at 2 µM) was pre-incubated with 100 nM of SARS-CoV-2 Mpro for 30 min at 30 °C, and then 10 µM FRET substrate was added to reaction mixture to initiate the reaction. The reaction was monitored for 2 h. The initial velocity was calculated by linear regression using the data points from the first 15 min of the reaction. The calculated initial velocity with each compound was normalized to DMSO control. The results are average ± standard deviation of two repeats.
Secondary screening of a focused library of calpain/cathepsin inhibitors and known viral 3CLpro inhibitors
Given the encouraging results from the primary screening, we then further characterized the four hits (24, 28, 29, and 43) in a combination of assays including dose-response titration, thermal shift binding assay (TSA), and counter screening assays with two other viral cysteine proteases, the enterovirus A71 (EV-A71) 2A and 3C proteases, both of which are cysteine proteases (Table 2). The HCV NS3–4A protease inhibitors boceprevir (28) and narlaprevir (29) inhibited Mpro with 50% inhibitory concentration (IC50) values of 4.13 and 4.73 µM, respectively (Table 2), more potent than simeprevir (24) (IC50 = 13.74 µM). Both compounds (28 and 29) also showed strong binding towards Mpro and shifted the melting temperature of the protein (ΔTm) by 6.67 °C and 5.18 °C, respectively, at 40 µM. Despite their potent inhibition against the HCV NS3–4A serine protease and the SARS-CoV-2 cysteine Mpro, boceprevir (28) and narlaprevir (29) did not inhibit the EV-A71 2A and 3C proteases (IC50 > 20 µM), suggesting that they are not non-specific cysteine protease inhibitors. The calpain inhibitor MG-132 (43) had an IC50 value of 3.90 µM against Mpro, and was not active against the EV-A71 2A and 3C proteases (IC50 > 20 µM). The binding of MG-132 (43) to Mpro was also confirmed in the TSA assay with a ΔTm of 4.02 °C.
Table 2.
Characterization of HCV and calpain/cathepsin protease inhibitors against SARS-CoV-2 Mpro using a consortium of secondary assaysa.
 | |
 |
aValue = means ± SE from three independent experiments.
bN.T. = not tested.
cThe IC50 of PSI (64) on SARS CoV-2 Mpro was calculated by end point reading of 1 h digestion, instead of the initial velocity.
In light of the promising results of the calpain inhibitor MG-132 (43), we then pursued testing for other calpain and cathepsin inhibitors that are commercially available (56–63) (Table 2). These compounds were not included in the initial library because they have not been advanced to clinical studies. Among this series of analogs, calpain inhibitor II (61) and XII (62) are the most potent Mpro inhibitors with IC50 values of 0.97 and 0.45 µM, respectively. Binding of compounds (61 and 62) to Mpro shifted the melting curve of the protein by 6.65 and 7.86 °C, respectively. Encouragingly, both compounds (61 and 62) did not inhibit the EV-A71 2A and 3C proteases (IC50 > 20 µM). Calpain inhibitor I (59) and MG-115 (60) also showed potent inhibition against Mpro with IC50 values of 8.60 and 3.14 µM, respectively. Calpeptin (56) and PSI (63) had moderate activity against Mpro with IC50 values of 10.69 and 10.38 µM, respectively. In contrast, calpain inhibitors III (57) and VI (58) were not active (IC50 > 20 µM).
We also included two well-known viral 3CL protease inhibitors GC-376 (64) and rupintrivir (65) in the secondary screening. GC-376 (64) is an investigational veterinary drug that is being developed for feline infectious peritonitis (FIP).13,14 GC-376 (64) was designed to target the viral 3CL protease and had potent antiviral activity against multiple viruses including MERS-CoV, FIPV, and norovirus.13,15 Rupintrivir (65) was developed as a rhinovirus antiviral by targeting the viral 3CL protease, but it was discontinued in clinical trials due to side effects.16 In our study, we found that GC-376 (64) was the most potent Mpro inhibitor with an IC50 value of 0.03 µM. It shifted the melting curve of Mpro by 18.30 °C upon binding. In contrast, rupintrivir (65) was not active against Mpro (IC50 > 20 µM). Previous report also showed that rupintrivir was not active against the SARS-CoV 3CLpro (Mpro) (IC50 > 100 µM).17 Both compounds (64 and 65) were inactive against the EV-A71 2A protease, but showed potent inhibition against the EV-A71 3C protease, which is consistent with previously reported results.15,18,19
When plotting the IC50 values (log scale) of the inhibitors against Mpro from the FRET enzymatic assay with the melting temperature shifts (ΔTm) from TSA (Fig. 3a), a linear correlation was observed, and the r2 of the linear regression fitting is 0.94. This suggests that there is a direct correlation between the enzymatic inhibition and protein binding: a more potent enzyme inhibitor also binds to the protein with higher affinity. The stabilization of the Mpro against thermal denaturation was also compound concentration dependent (Fig. 3b).
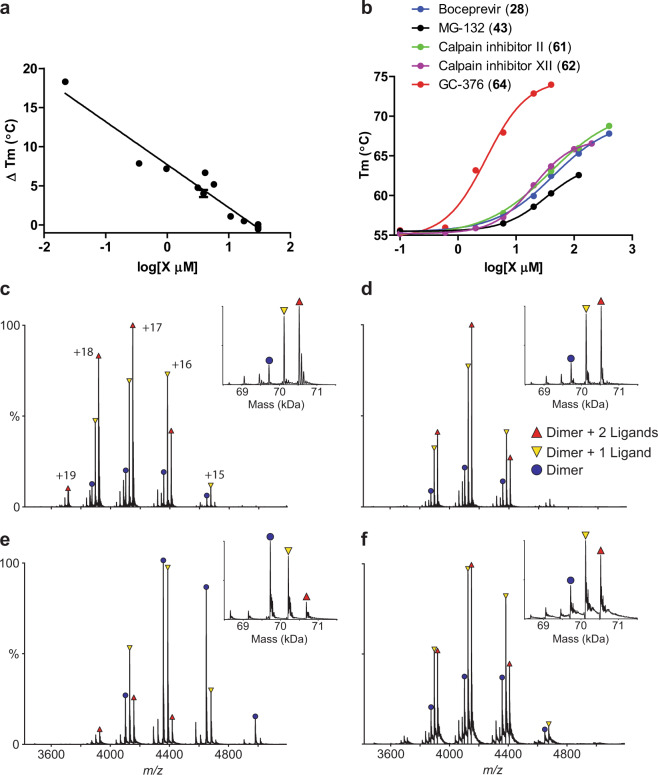
Fig. 3. Binding of inhibitors to SARS-CoV-2 Mpro using thermal shift binding assay and native mass spectrometry.
a Correlation of inhibition efficacy (IC50) with ΔTm from thermal shift binding assay. Data in Table 2 were used for the plot. The r2 of fitting is 0.94. b Dose-dependent melting temperature (Tm) shift. Native MS reveals binding of SARS-CoV-2 Mpro to GC-376 (64) (c), calpain inhibitor II (61) (d), calpain inhibitor XII (62) (e), and boceprevir (28) (f). All ligand concentrations are 12.5 µM except (e), which is 25 µM. Peaks are annotated for dimer (blue circle), dimer with one bound ligand (yellow down triangle), and dimer with two bound ligands (red up triangle). Other minor signals are truncated dimers, which bind ligands at the same ratios. Charge states are annotated in c, and insets show the deconvolved zero-charge mass distribution.
The binding of the four most potent inhibitors boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64) to SARS-CoV-2 Mpro was further characterized by native mass spectrometry (MS) (Fig. 3c–f). Native MS analysis showed that Mpro formed a dimer complex with a mass of 69,722 Da, indicating that the extra N-terminal methionine in our Mpro construct was cleaved, yielding the native Mpro N-terminus with a serine residue (Supplementary information, Fig. S1). A small amount of protomer and dimer with a single C-terminal truncation of the His-tag was also observed, but the intact dimer was the predominant signal (Supplementary information, Fig. S1). Addition of all four ligands tested, boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64), showed binding of up to two ligands per dimer (Fig. 3c–f), suggesting a binding stoichiometry of one drug per protomer.
Mechanism of action of hits
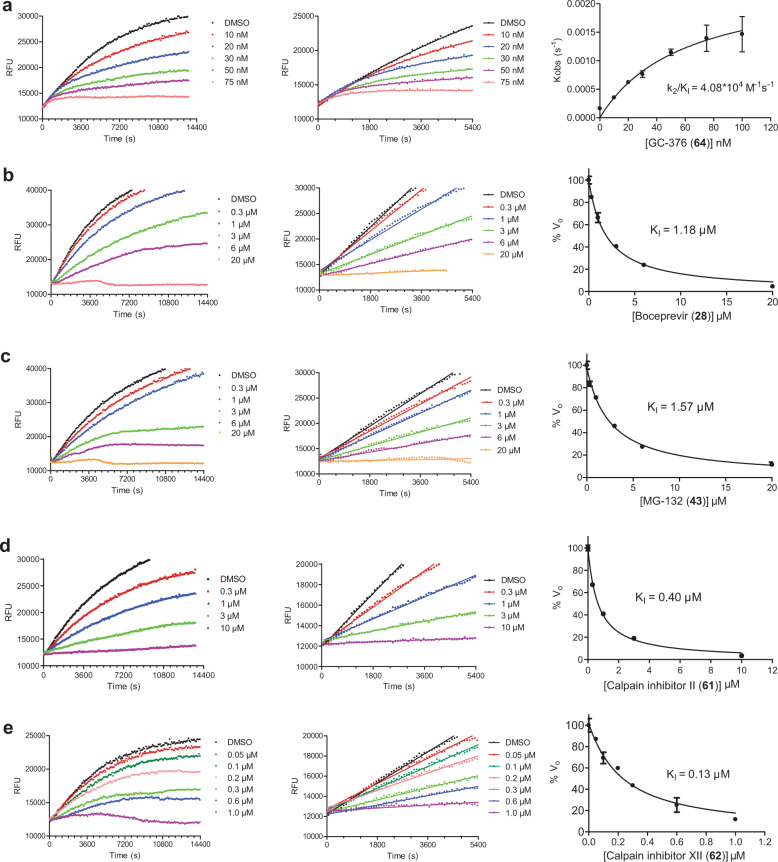
To elucidate the mechanism of action of hits against SARS-CoV-2 Mpro, we focus on five most potent compounds prioritized from the primary and secondary screenings including boceprevir (28), MG-132 (43), calpain inhibitor II (61), calpain inhibitor XII (62), and GC-376 (64). For this, we performed enzyme kinetic studies with different concentrations of inhibitors (Fig. 4). A biphasic enzymatic progression curve in the presence but not absence of inhibitor is typically a hallmark for a slow covalent binding inhibitor. In Fig. 4, the left column shows the progression curves up to 4 h. Biphasic progression curves were observed for all 5 inhibitors at high drug concentrations. Significant substrate depletion was observed when the proteolytic reaction proceeded beyond 90 min. We therefore chose the first 90 min of the progression curves for curve fitting (Fig. 4, middle column). We fit the progression curves in the presence of different concentrations of GC-376 (64) with the two-step Morrison equation (Eq. (3) in “Materials and methods” section). GC-376 (64) binds to SARS-CoV-2 Mpro with an equilibrium dissociation constant for the inhibitor (KI) of 59.9 ± 21.7 nM in the first step. After initial binding, a covalent bond is formed at a slower velocity between GC-376 (64) and Mpro with the second reaction rate constant (k2) being 0.00245 ± 0.00047 s−1, resulting in an overall k2/KI value of 4.08 × 104 M−1 s−1 (Fig. 4a). However, when we tried to fit the proteolytic progression curves for boceprevir (28), MG-132 (43), calpain inhibitors II (61) and XII (62) using the same two-step reaction mechanism, we could not obtain accurate values for the second rate constant k2. This is presumably due to significant substrate depletion before the equilibrium between EI and EI*, leading to very small values of k2. Accordingly, for these four inhibitors (28, 43, 61, and 62), only the dissociation constant KI values from the first step were determined (Fig. 4b–e). The inhibition constants (KI) for boceprevir (28), MG-132 (43), calpain inhibitors II (61) and XII (62) are 1.18 ± 0.10 µM, 1.57 ± 0.13 µM, 0.40 ± 0.02 µM, and 0.13 ± 0.02 µM, respectively.
Fig. 4. Proteolytic reaction progression curves of Mpro in the presence or the absence of compounds.
a–e In the kinetic studies, 5 nM Mpro was added to a solution containing various concentrations of protease inhibitors and 20 µM FRET substrate to initiate the reaction. The reaction was then monitored for 4 h. The left column shows the reaction progression up to 4 h; middle column shows the progression curves for the first 90 min, which were used for curve fitting to generate the plot shown in the right column. Detailed methods were described in “Materials and methods” section. GC-376 (64) (a); boceprevir (28) (b); MG-132 (43) (c); calpain inhibitor II (61) (d); calpain inhibitor XII (62) (e).
Cellular antiviral activity and cytotoxicity of hits
To test the hypothesis that inhibiting the enzymatic activity of Mpro will lead to the inhibition of SARS-CoV-2 viral replication, we performed cellular antiviral assays for the five promising hits (64, 28, 43, 61, and 62) against SARS-CoV-2. For this, we first tested the cellular cytotoxicity of these compounds in multiple cell lines (Supplementary information, Table S1). GC-376 (64), boceprevir (28), and calpain inhibitor II (61) were well tolerated and had CC50 values of over 100 µM for all the cell lines tested. MG-132 (43) was cytotoxic to all the cells with CC50 values less than 1 µM except A549 cells. Calpain inhibitor XII (62) had acceptable cellular cytotoxicity with CC50 values above 50 µM for all the cell lines tested (Supplementary information, Table S1).
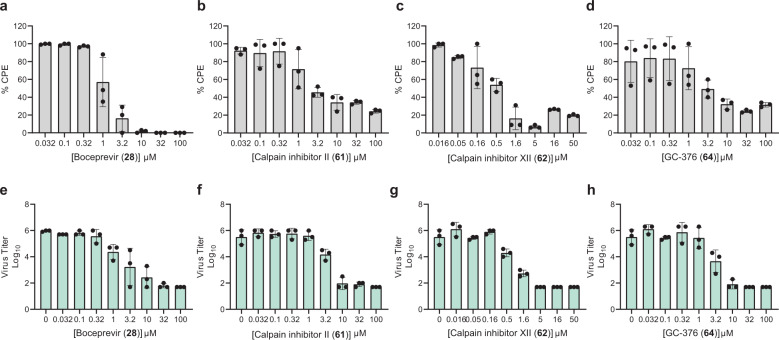
Next, we chose four compounds boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64) for the antiviral assay with infectious SARS-CoV-2. MG-132 (43) was not included due to its cytotoxicity. Gratifyingly, all four compounds showed potent antiviral activity against SARS-CoV-2 in the primary viral cytopathic effect (CPE) assay with 50% effective concentration (EC50) values ranging from 0.49 to 3.37 µM (Fig. 5a–d and Table 3). Their antiviral activity was further confirmed in the secondary viral yield reduction (VYR) assay (Fig. 5e–h and Table 3). The most potent compound was calpain inhibitor XII (62), which showed an EC50 of 0.49 µM in the primary CPE assay and an EC50 of 0.78 µM in the secondary VYR assay (Table 3; Supplementary information, Fig. S2). In comparison, remdesivir was reported to inhibit SARS-CoV-2 in the VYR assay with an EC50 of 0.77 µM.20 None of the compounds inhibited the unrelated influenza virus A/California/07/2009 (H1N1) virus (EC50 > 20 µM) (Supplementary information, Table S1), suggesting that the antiviral activity of the four compounds (boceprevir, calpain inhibitors II, XII, and GC-376) against SARS-CoV-2 is specific. In comparison with recently reported SARS-CoV-2 Mpro inhibitors (Table 3), the hits identified herein represent some of the most potent and selective drug candidates with broad chemical diversity.
Fig. 5. Antiviral potency of boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64), in CPE and VYR assays.
a–d Primary CPE assay results of boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64). e–h Secondary VYR assay results of boceprevir (28), calpain inhibitors II (61), XII (62), and GC-376 (64). Each dot represents the average of three repeats from one experiment, and each assay was repeated in three different experiments. The curve fittings were shown in Supplementary information, Fig. S2.
Table 3.
Antiviral activity of hits against SARS-CoV-2 and the comparison with recently reported Mpro inhibitors.
 |
Complex crystal structure of SARS-CoV-2 Mpro with GC-376 (64)
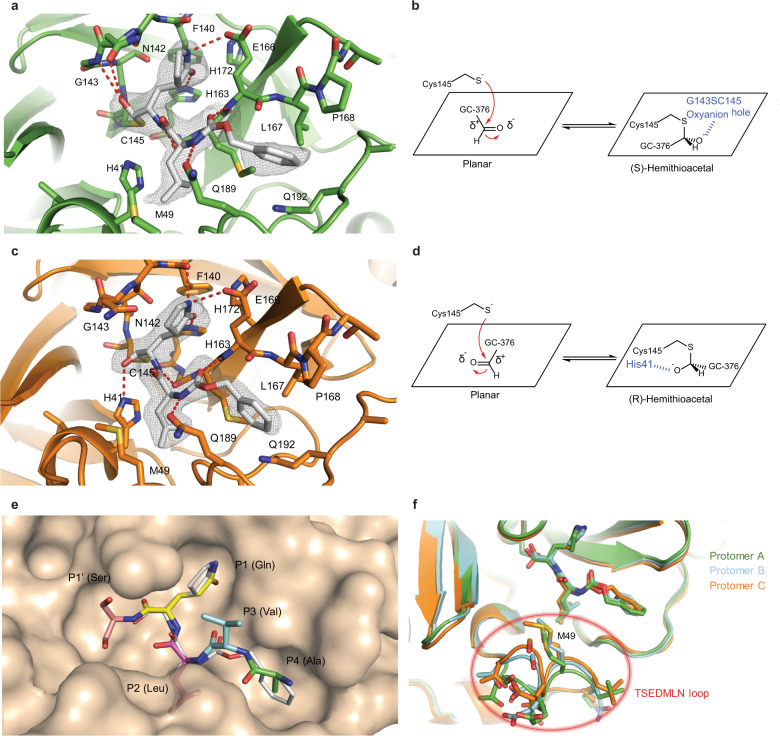
The crystal structure of the SARS-CoV-2 Mpro in complex with GC-376 (64) was solved in the P3221 space group at 2.15 Å resolution (PDB: 6WTT) (Supplementary information, Table S2). There are three protomers per asymmetric unit (ASU) with two constituting a biological dimer and the third forming a dimer with a crystallographic symmetry related neighboring protomers (Supplementary information, Fig. S3). The presence of three protomers in our crystal structure allowed us to capture different binding configurations of GC-376 (64) (Fig. 6), a unique feature that was not observed in previous X-ray crystal structures.8,10,11 The pairwise r.m.s.d among the protomer backbone Cα atoms ranges from 0.435 to 0.564 Å. Previously, SARS-CoV Mpro and SARS-CoV-2 Mpro crystal structures have been solved most frequently as one protomer per ASU, and occasionally a dimer.8,10,11,21,22 In its native state, Mpro requires dimerization to become catalytically active23,24, which is supported by our native MS data (Supplementary information, Fig. S1). In our crystal structures, all three protomers appear catalytically competent, with the third protomer activated by the N-finger from an adjacent ASU. The well-defined electron density also clearly shows a serine at the protease N-terminus, again indicating that the first methionine in the protease construct was cleaved (Supplementary information, Fig. S3b).
Fig. 6. Molecular recognition of GC-376 (64) by SARS-CoV-2 Mpro.
Complex of SARS-CoV-2 Mpro and GC-376 (64) with protomer A (a, b) and protomer C (c, d). Unbiased Fo–Fc map, shown in gray, is contoured at 2σ. Hydrogen bonds are shown as red dashed lines. e Surface representation of SARS-CoV-2 Mpro + GC-376 (64) (white) superimposed with the SARS-CoV Mpro natural, N-terminal substrate (PDB ID: 2Q6G, with residues P1′–P4 in different colors). The SARS-CoV-2 Mpro cleaves between the P1′ and P1 residues. f Superimposition of the three protomers in the asymmetric subunit of SARS-CoV-2 Mpro with GC-376 (64). Significant conformational flexibility is observed, particularly in the TSEDMLN loop.
GC-376 (64) forms an extensive network of hydrogen bonds with the active site while also exhibiting excellent geometric complementarity (Fig. 6). These interactions are coupled with the thermodynamic payoff of covalent adduct formation between the aldehyde bisulfite warhead and Cys145, making GC-376 (64) one of the most potent SARS-CoV-2 Mpro inhibitors in vitro with an IC50 of 0.030 ± 0.008 µM (Table 3). Along with other known Mpro inhibitors (N3, 13b, 11a and 11b) (Table 3), GC-376 (64) mimics the peptide substrate that is cleaved by this enzyme (Fig. 6e).8,22,25,26 The glutamine surrogate γ-lactam ring is a cyclized derivative of the P1 glutamine side chain that normally occupies the S1 site; here it forms hydrogen bonds with the His163 and Glu166 side chains and the main chain of Phe140 (Fig. 6a, c). An amide bond connects the γ-lactam side chain to an isobutyl moiety that embeds itself in the hydrophobic S2 site formed by His41, Met49, and Met169. Normally, this S2 site in SARS-CoV-2 Mpro can accommodate a variety of hydrophobic substitutions such as isobutyl in GC-376 (64) and N3), cyclopropyl in (13b), cyclohexyl in (11a), and 3-fluorophenyl in (11b) (Table 3).27,28 A carbamate bond in GC-376 (64), which forms hydrogen bonds with the main chain of Glu166 and the side chain of Gln189, connects the P2 isobutyl group to a phenylmethyl ester that interacts with the aliphatic S4 site. Compared with previous inhibitors, the phenylmethyl ester of GC-376 (64) exhibits high complementarity with the S4 site, and the extensive non-polar interactions may contribute significantly to the potency of this compound (Fig. 6e).
Three copies of GC-376 (64) were found in the crystal structure, one in each protomer active site (Fig. 6a, c; Supplementary information, Fig. S4). The configurations of GC-376 (64) were consistent in protomers A and B, where the thioacetal hydroxide is positioned in the “oxyanion hole” formed by the backbone amides of Gly143, Ser144, and Cys145 (Fig. 6a; Supplementary information, Fig. S4), resulting in the (S)-configuration. It is noted that aldehydes (11a and 11b) also bind the active site of SARS-CoV-2 Mpro in the (S)-configuration (PDB: 6M0K and 6LZE) (Supplementary information, Fig. S5a).11 In protomer C, however, the same hydroxide group orients outwards from the oxyanion hole, forming hydrogen bonds with His41 (Fig. 6c), which gives the (R)-configuration. This (R)-configuration is consistent with the binding mode of α-ketoamide (13b) in the active site of SARS-CoV-2 Mpro (PDB: 6Y2F) (Supplementary information, Fig. S5b).8 These two unique configurations R and S might be a result of the Cys145 thiol nucleophilic attacking the aldehyde of GC-376 (64) from two different faces (Fig. 6b, d). The fact that GC-376 (64) can adopt two different configurations R and S upon binding to the active site might explain its high binding affinity towards the target.
An additional difference between the configurations of GC-376 (64) in protomers A, B and C is observed in the orientation of the phenylmethyl ester. In protomer C, the CH2 of the phenylmethyl points toward the main chain of Leu167 in a “cis” conformation (Fig. 6c), whereas in protomers A and B this same CH2 points downwards in a “trans” conformation (Fig. 6a; Supplementary information, Fig. S4). Consequently, this influences the rotameric configuration of the Leu167 isobutyl moiety, where a rotational adjustment of 180° occurs at its “β” carbon. Furthermore, large rearrangements are observed in the flexible loop that consists of residues 45–51 (TSEDMLN in SARS-CoV-2 Mpro) and forms the S2 and S3′ subsites (Fig. 6f), explaining the broad substrate scope in the P2 site (Table 3). The loop conformations in protomers B and C may be influenced by crystal packing interactions with protomers from adjacent ASUs and resemble the conformations in previously determined structures.10,11 Meanwhile, the conformation of protomer A is less restrained and exhibits the most significant conformational divergence. The different loop conformations offer a glimpse of the protein plasticity that allows Mpro to accommodate peptides with differing amino acid composition, and underscores the importance of considering this flexibility when analyzing and modeling protein-ligand interactions for Mpro.
Discussion
Coronaviruses have caused three epidemics/pandemics in the past 20 years including SARS, MERS, and COVID-19. With the ongoing pandemic of COVID-19, scientists and researchers around the globe are racing to find effective vaccines and antiviral drugs.6 The viral polymerase inhibitor remdesivir holds the greatest promise and it is currently being evaluated in several clinical trials.29,30 Remdesivir was recently granted emergency use authorization by FDA. The HIV drug combination lopinavir and ritonavir recently failed in a clinical trial for COVID-19 as no significant therapeutic efficacy was observed.31 To address this unmet medical need, we initiated a drug repurposing screening to identify potent inhibitors against the SARS-CoV-2 Mpro from a collection of FDA-approved protease inhibitors. The Mpro has been shown to be a validated antiviral drug target for SARS-CoV and MERS-CoV.32 As the SARS-CoV-2 Mpro shares a high sequence similarity with SARS-CoV and to a less extent with MERS-CoV, we reasoned that inhibiting the enzymatic activity of SARS-CoV-2 Mpro will similarly prevent viral replication.8,10
Noticeable findings from our study include: (1) Boceprevir (28), an FDA-approved HCV drug, inhibits the enzymatic activity of Mpro with IC50 of 4.13 µM, and has an EC50 of 1.90 µM against the SARS-CoV-2 virus in the cellular viral CPE assay. The therapeutic potential of boceprevir (28) should be further evaluated in relevant animal models and human clinical trials. Since boceprevir (28) is an FDA-approved drug, the dose, toxicity, formulation and pharmacokinetic properties are already known, which will greatly speed up the design of follow-up studies; (2) GC-376 (64), an investigational veterinary drug, showed promising antiviral activity against the SARS-CoV-2 virus (EC50 = 3.37 µM). It has the highest enzymatic inhibition against the Mpro with an IC50 value of 0.03 µM. This compound has promising in vivo efficacy in treating cats infected with FIP, and has favorable in vivo pharmacokinetic properties. Therefore, GC-376 (64) is ready to be tested in relevant animal models of SARS-CoV-2 when available. Importantly, the X-ray crystal structure of SARS-CoV-2 Mpro in complex with GC-376 (64) provides a molecular explanation of the high binding affinity of aldehyde-containing compounds as they can adopt two configurations R and S. The conformational flexibility at the TSEDMLN loop explains the broad substrate scope at the P2 position of Mpro inhibitors; (3) Three calpain/cathepsin inhibitors, MG-132 (43), calpain inhibitors II (61) and XII (62), are potent inhibitors of Mpro with single-digit to submicromolar efficacy in the enzymatic assay. Calpain inhibitors II (61) and XII (62) also inhibit SARS-CoV-2 in the CPE assay with EC50 values of 2.07 and 0.49 µM, respectively. This result suggests that calpain/cathepsin inhibitors are rich sources of drug candidates for SARS-CoV-2. Indeed, previous studies have shown that calpain II and cathepsin L are required for the proteolytic processing of the coronavirus S protein, a step that is essential for the viral fusion and genome release during the early stage of viral replication.33 Calpain and cathepsin inhibitors such as MDL28170 (calpain inhibitor III),33 MG-132,34 calpain inhibitor VI35 have been shown to inhibit SARS-CoV replication in cell culture. Other than the increased potency of targeting both Mpro and calpain/cathepsin, an additional benefit of such dual inhibitors might be their high genetic barrier to drug resistance. A significant number of calpain/cathepsin inhibitors have been developed over the years for various diseases including cancer, neurodegeneration disease, kidney diseases, and ischemia/reperfusion injury.36 Given our promising results of calpain inhibitors II (61) and XII (62) in inhibiting the SARS-CoV-2 Mpro and their potent antiviral activity in cell culture, it might be worthwhile to repurpose them as antivirals for SARS-CoV-2.
All potent SARS-CoV-2 Mpro inhibitors contain reactive warheads such as α-ketoamide (boceprevir (28), calpain inhibitor XII (62)) or aldehyde (MG-132 (43), calpain inhibitor II (61)) or aldehyde prodrug, the bisulfite (GC-376 (64)). This result suggests that reactive warheads might be essential for SARS-CoV-2 Mpro inhibition. The compounds identified in this study represent some of the most potent and selective hits reported so far, and have similar activity to the recently reported SARS-CoV-2 Mpro inhibitors (ebselen, N3, and 13b) (Table 3). Calpain inhibitor XII (62) had similar potency to the recently disclosed compounds (11a and 11b) (Table 3).11 Notably, boceprevir (28), calpain inhibitor II (61) and XII (62) have different chemical scaffolds than the substrate mimetics GC-376 (64), (N3, 13b, 11a, and 11b), therefore providing new opportunities for designing more potent and selective SARS-CoV-2 Mpro inhibitors.
Aside from the above positive results, we also showed that ritonavir (9) and lopinavir (10) failed to inhibit the SARS-CoV-2 Mpro (IC50 > 20 µM, Fig. 2), which might explain why they lack efficacy in clinical trials for COVID-19.31 Camostat (39) was recently proposed to inhibit SARS-CoV-2 entry through inhibiting the host TMPRSS2, a host serine protease that is important for viral S protein priming.37 In our study, we found that camostat (39) has no inhibition against the SARS-CoV-2 Mpro (IC50 > 20 µM).
In summary, this study identified several potent SARS-CoV-2 Mpro inhibitors with potent enzymatic inhibition as well as cellular antiviral activity. Further development based on these hits might lead to clinically useful COVID-19 antivirals. They can be used either alone or in combination with polymerase inhibitors such as remdesivir as a means to achieve potential synergistic antiviral effect as well as to suppress drug resistance.
Materials and methods
Cell lines and viruses
Human rhabdomyosarcoma RD, A549, MDCK, Caco-2, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM); BEAS2B and HCT-8 cells were maintained in RPMI 1640 medium. Both medium was supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin antibiotics. Cells were kept at 37 °C in a 5% CO2 atmosphere. The USA-WA1/2020 strain of SARS-CoV-2 was obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA).
Protein expression and purification
SARS-CoV-2 main protease (Mpro or 3CL) gene from strain BetaCoV/Wuhan/WIV04/2019 was ordered from GenScript (Piscataway, NJ, USA) in the pET29a(+) vector with E. coli codon optimization. pET29a(+) plasmids with SARS-CoV-2 Mpro was transformed into competent E. coli BL21(DE3) cells, and a single colony was picked to inoculate 10 mL of LB supplemented with 50 g/mL kanamycin and grown at 37 °C with shaking at 250 rpm. The 10-mL inoculum was added to 1 L of LB with 50 g/mL kanamycin and grown to an optical density at 600 nm of 0.8, and then induced using 1.0 mM IPTG. Induced cultures were incubated at 37 °C for an additional 3 h and then harvested, resuspended in lysis buffer (25 mM Tris, pH 7.5, 750 mM NaCl, 2 mM DTT with 0.5 mg/mL lysozyme, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.02 mg/mL DNase I), and lysed by alternating sonication and French press cycles. The cell debris were removed by centrifugation at 12,000× g for 45 min (20% amplitude, 1 s on/1 s off). The supernatant was incubated with Ni-NTA resin for over 2 h at 4 °C on a rotator. The Ni-NTA resin was thoroughly washed with 30 mM imidazole in wash buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 2 mM DTT); and eluted with 100 mM imidazole in elution buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 2 mM DTT). The imidazole was removed via dialysis or on a 10,000-molecular-weight-cutoff centrifugal concentrator spin column. The purity of the protein was confirmed with SDS-PAGE. The protein concentration was determined via 260 nm absorbance with ε 32890. EV-A71 2Apro and 3Cpro were expressed in the pET28b(+) vector as previously described.18,38,39
The protein sequence of the SARS-CoV-2 Mpro used in the enzymatic assay and X-ray crystallization is listed below:
SGFRKMAFPS GKVEGCMVQV TCGTTTLNGL WLDDVVYCPR HVICTSEDML NPNYEDLLIR KSNHNFLVQA GNVQLRVIGH SMQNCVLKLK VDTANPKTPK YKFVRIQPGQ TFSVLACYNG SPSGVYQCAM RPNFTIKGSF LNGSCGSVGF NIDYDCVSFC YMHHMELPTG VHAGTDLEGN FYGPFVDRQT AQAAGTDTTI TVNVLAWLYA AVINGDRWFL NRFTTTLNDF NLVAMKYNYE PLTQDHVDIL GPLSAQTGIA VLDMCASLKE LLQNGMNGRT ILGSALLEDE FTPFDVVRQC SGVTFQ LEHHHHHH.
Peptide synthesis
The SARS-CoV-2 Mpro FRET substrate Dabcyl-KTSAVLQ/SGFRKME (Edans) was synthesized by solid-phase synthesis through iterative cycles of coupling and deprotection using the previously optimized procedure.40 Specifically, chemmatrix rink-amide resin was used. Typical coupling condition was 5 equiv of amino acid, 5 equiv of HATU, and 10 equiv of DIEA in DMF for 5 min at 80 °C. For deprotection, 5% piperazine plus 0.1 M HOBt were used and the mixture was heated at 80 °C for 5 min. The peptide was cleaved from the resin using 95% TFA, 2.5% Tris and 2.5% H2O, and the crude peptide was precipitated from ether after removal of TFA. The final peptide was purified by preparative HPLC. The purify and identity of the peptide were confirmed by analytical HPLC (> 98% purity) and mass spectrometry. [M + 3]3+ was calculated as 694.15 and detected as 694.90; [M + 4]4+ was calculated as 520.86 and detected as 521.35.
Edans standard curve
Edans standard curve was generated as described below: 200 nM SARS-CoV-2 Mpro was incubated with varying concentrations of FRET substrate (0.5–40 µM) and the reaction progress was monitored until the fluorescence signals reached plateau, at which point we deemed all the FRET substrate was digested by Mpro. The endpoint fluorescence signal was plotted against FRET substrate concentration with a linear regression function in Prism 5.
Native mass spectrometry
Prior to analysis, the protein was buffer exchanged into 0.2 M ammonium acetate (pH 6.8) and diluted to 10 μM. DTT was dissolved in water and prepared at a 400 mM stock. Each ligand was dissolved in ethanol and diluted to 10× stock concentrations. The final mixture was prepared by adding 4 μL protein, 0.5 μL DTT stock, and 0.5 μL ligand stock for final concentration of 4 mM DTT and 8 μM protein. Final ligand concentrations were used as annotated. The mixtures were then incubated for 10 min at room temperature prior to analysis. Each sample was mixed and analyzed in triplicate.
Native MS was performed using a Q-Exactive HF quadrupole-Orbitrap mass spectrometer with the Ultra-High Mass Range research modifications (Thermo Fisher Scientific). Samples were ionized using nano-electrospray ionization in positive ion mode using 1.0 kV capillary voltage at a 150 °C capillary temperature. The samples were all analyzed with a 1000–25,000 m/z range, the resolution set to 30,000, and a trapping gas pressure set to 3. Source fragmentation (10–50 V) was applied to all samples to aid in desolvation. Data were deconvolved and analyzed with UniDec.41
Enzymatic assays
For reaction condition optimization, 200 µM SARS-CoV-2 Mpro was used. pH 6.0 buffer contains 20 mM MES, pH 6.0, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT and 20% glycerol; pH 6.5 buffer contains 20 mM HEPES, pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT and 20% glycerol; pH 7.0 buffer contains 20 mM HEPES, pH 7.0, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT and 20% glycerol. Upon addition of 20 µM FRET substrate, the reaction progress was monitored for 1 h. The first 15 min of reaction was used to calculate initial velocity via linear regression in Prism 5. Mpro displays the highest proteolytic activity in pH 6.5 buffer. All the following enzymatic assays were carried out in pH 6.5 buffer.
For the measurements of Km/Vmax, screening of the protease inhibitor library, as well as IC50 measurements, proteolytic reaction with 100 nM Mpro in 100 µL of pH 6.5 reaction buffer was carried out at 30 °C in a Cytation 5 imaging reader (Thermo Fisher Scientific) with filters for excitation at 360/40 nm and emission at 460/40 nm. Reactions were monitored every 90 s. For Km/Vmax measurements, a FRET substrate concentration ranging from 0 to 200 µM was applied. The initial velocity of the proteolytic activity was calculated by linear regression for the first 15 min of the kinetic progress curves. The initial velocity was plotted against the FRET concentration with the classic Michaelis–Menten equation in Prism 5 software. For the screening of protease inhibitor library and IC50 measurements, 100 nM Mpro was incubated with protease inhibitor at 30 °C for 30 min in reaction buffer, then the reaction was initiated by adding 10 µM FRET substrate, the reaction was monitored for 1 h, and the initial velocity was calculated for the first 15 min by linear regression. The IC50 was calculated by plotting the initial velocity against various concentrations of protease inhibitors by use of a dose-response curve in Prism 5 software. Proteolytic reaction progress curve kinetics measurements with GC376, MG132, boceprevir, calpain inhibitor II, and calpain inhibitor XII used for curve fitting, were carried out as follows: 5 nM Mpro protein was added to 20 µM FRET substrate with various concentrations of testing inhibitor in 200 µL of reaction buffer at 30 °C to initiate the proteolytic reaction. The reaction was monitored for 4 h. The progress curves were fit to a slow binding Morrison equation (Eq. (3)) as described previously18,42:
| 1 |
| 2 |
| 3 |
| 4 |
where P(t) is the fluorescence signal at time t, P0 is the background signal at time 0, V0, Vs, and k represent, respectively, the initial velocity, the final steady-state velocity and the apparent first-order rate constant for the establishment of the equilibrium between EI and EI*.42 k2/KI is commonly used to evaluate the efficacy for covalent inhibitor. We observed substrate depletion when proteolytic reactions progress longer than 90 min; therefore only first 90 min of the progress curves were used in the curve fitting (Fig. 6, middle column). In this study, we could not accurately determine the k2 for the protease inhibitors: calpain inhibitor II, MG132, boceprevir, and calpain inhibitor XII, due to the very slow k2 in these cases: significant substrate depletion before the establishment of the equilibrium between EI and EI*. In these cases, KI was determined with Morrison equation in Prism 5.
Differential scanning fluorimetry
The binding of protease inhibitors to Mpro was monitored by differential scanning fluorimetry (DSF) using a Thermo Fisher QuantStudioTM 5 Real-Time PCR System. TSA plates were prepared by mixing Mpro protein (final concentration of 3 μM) with inhibitors, and incubated at 30 °C for 30 min. 1× SYPRO orange (Thermo Fisher Scientific) were added and the fluorescence of the plates were taken under a temperature gradient ranging from 20 to 90 °C (incremental steps of 0.05 °C/s). The melting temperature (Tm) was calculated as the mid-log of the transition phase from the native to the denatured protein using a Boltzmann model (Protein Thermal Shift Software v1.3). Thermal shift that was represented as ΔTm was calculated by subtracting reference melting temperature of proteins in DMSO from the Tm in the presence of compound.
Cytotoxicity measurement
A549, MDCK, HCT-8, Caco-2, Vero, and BEAS2B cells for cytotoxicity CPE assays were seeded and grown overnight at 37 °C in a 5% CO2 atmosphere to ∼90% confluence on the next day. Cells were washed with phosphate-buffered saline (PBS) and 200 µL DMEM containing 2% FBS and 1% penicillin–streptomycin, and various concentrations of protease inhibitors were added to each well. 48 h after addition of the protease inhibitors, cells were stained with 66 μg/mL neutral red for 2 h, and neutral red uptake was measured at an absorbance at 540 nm using a Multiskan FC microplate photometer (Thermo Fisher Scientific). The CC50 values were calculated from best-fit dose-response curves using GraphPad Prism 5 software.
SARS-CoV-2 CPE assay
Antiviral activities of test compounds were determined in nearly confluent cultures of Vero 76 cells. The assays were performed in 96-well Corning microplates. Cells were infected with approximately 60-fold of 50% cell culture infectious doses (CCID50) of SARS-CoV-2 and 50% effective concentrations (EC50) were calculated based on virus-induced CPE quantified by neutral red dye uptake after 5 days of incubation. USA-WA1/2020 strain of SARS-CoV-2 obtained from the WRCEVA was used for both CPE and VYR assays. Three microwells at each concentration of compound were infected. Two uninfected microwells served as toxicity controls. Cells were stained for viability with neutral red (0.11% final concentration) for 2 h. Excess dye was rinsed from the cells with PBS. The absorbed dye was eluted from the cells with 0.1 mL of 50% Sörensen’s citrate buffer (pH 4.2)-50% ethanol. Plates were read for optical density determination at 540 nm. Readings were converted to the percentage of the results for the uninfected control using an Excel spreadsheet developed for this purpose. EC50 values were determined by plotting percent CPE versus log10 inhibitor concentration. Toxicity at each concentration was determined in uninfected wells in the same microplates by measuring dye uptake.
SARS-CoV-2 VYR assay
VYR assays were conducted by first replicating the viruses in the presence of test compound. Supernatant was harvested 3 days post infection from each concentration of test compound and the virus yield was determined by endpoint dilution method. Briefly, supernatant virus was serially diluted in log10 increments and then plated onto quadruplicate wells of 96-well plates seeded with Vero 76 cells. The presence or absence of CPE for determining a viral endpoint was evaluated by microscopic examination of cells 6 days post infection. From these data, 90% virus inhibitory concentrations (EC90) were determined by regression analysis.
Influenza A virus A/California/07/2009 (H1N1) plaque reduction assay
The plaque assay was performed according to previously published procedures.43
Mpro crystallization and structure determination
10 mg/mL of SARS-CoV-2 Mpro was incubated with 2 mM GC376 at 4 °C overnight. The protein was diluted to 2.5 mg/mL the following day in protein buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 4 mM DTT). Since GC3760 is water soluble, no precipitation was observed, and centrifugation was not necessary. Crystals were grown by mixing 2 μL of the protein solution with 1 μL of the precipitant solution (15% PEG 2 K, 10% 1,6-hexanediol, and 0.2 M NaCl) in a hanging-drop vapor-diffusion apparatus. Crystals were cryoprotected by transferring to a cryoprotectant solution (20% PEG 2 K, 10% 1,6-hexanediol, 20% glycerol) and flash-frozen in liquid nitrogen.
X-ray diffraction data for the SARS-CoV-2 Mpro–GC376 complex structure was collected on the SBC 19-ID beamline at the Advanced Photon Source (APS) in Argonne, IL, and processed with the HKL3000 software suite.44 The CCP4 versions of MOLREP was used for molecular replacement using a previously solved SARS-CoV-2 Mpro structure (PDB ID: 5RGG) as a reference model.45 Rigid and restrained refinements were performed using REFMAC and model building was performed with COOT.46,47 Protein structure figures were made using PyMOL (Schrödinger, LLC).
Supplementary information
Acknowledgements
This research was partially supported by the National Institutes of Health (NIH) (Grant AI147325) and the Arizona Biomedical Research Centre Young Investigator grant (ADHS18-198859) to J.W., B.H. and B.T. thanks for the support from the Respiratory Diseases Branch, National Institute of Allergy and Infectious Diseases, NIH, USA (Contract N01-AI-30048). J.A.T. and M.T.M. were funded by the National Institute of General Medical Sciences, NIH (Grant R35 GM128624 to M.T.M.). We thank Michael Kemp for assistance with crystallization and X-ray diffraction data collection. We also thank the staff members of the Advanced Photon Source of Argonne National Laboratory, particularly those at the Structural Biology Center (SBC), with X-ray diffraction data collection. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Author contributions
J.W. and C.M. conceived and designed the study; C.M. expressed the Mpro with the assistance of T.S.; C.M. performed the primary screening, secondary IC50 determination, thermal shift binding assay, and enzymatic kinetic studies; M.D.S. carried out Mpro crystallization and structure determination with the assistance of X.Z., and analyzed the data with Y.C.; B.H. and B.T. performed the SARS-CoV-2 CPE and VYR assays; J.A.T. performed the native mass spectrometry experiments with the guidance from M.T.M.; Y.H. performed the plaque reduction assay with influenza A/California/07/2009 (H1N1) virus; J.W. and Y.C. secured funding and supervised the study; J.W., Y.C., and C.M. wrote the paper with the input from others.
Data availability
The structure for SARS-CoV-2 Mpro has been deposited in the Protein Data Bank with accession number 6WTT.
Competing interests
J.W. and C.M. are inventors of a pending patent that claims the use of the identified compounds for COVID-19.
Contributor Information
Yu Chen, Email: ychen1@usf.edu.
Jun Wang, Email: junwang@pharmacy.arizona.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0356-z.
References
- 1.Mahase E. Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368:m641. doi: 10.1136/bmj.m641. [DOI] [PubMed] [Google Scholar]
- 2.Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu A, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020). [DOI] [PMC free article] [PubMed]
- 5.Tang B, et al. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect. Dis. Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 7.Xia S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 10.Jin Z, et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 11.Dai W, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingfield PT. N-terminal methionine processing. Curr. Protoc. Protein Sci. 2017;88:6.14.1–6.14.3. doi: 10.1002/cpps.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen NC, et al. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 2018;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12:e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, et al. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayden FG, et al. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shie JJ, et al. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic alpha,beta-unsaturated esters. Bioorg. Med. Chem. 2005;13:5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musharrafieh R, et al. Validating enterovirus D68-2A(pro) as an antiviral drug target and the discovery of telaprevir as a potent D68-2A(pro) inhibitor. J. Virol. 2019;93:e0221-18. doi: 10.1128/JVI.02221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo CJ, et al. Design, synthesis, and evaluation of 3C protease inhibitors as anti-enterovirus 71 agents. Bioorg. Med. Chem. 2008;16:7388–7398. doi: 10.1016/j.bmc.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muramatsu T, et al. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl Acad. Sci. USA. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, et al. Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J. Biol. Chem. 2010;285:28134–28140. doi: 10.1074/jbc.M109.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand K, et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue X, et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, et al. Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 27.Chuck CP, et al. Profiling of substrate specificity of SARS-CoV 3CL. PLoS ONE. 2010;5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegyi A, Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 29.Grein J, et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holshue ML, et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao B, et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider M, et al. Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. J. Virol. 2012;86:10112–10122. doi: 10.1128/JVI.01001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnard DL, et al. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-D-N4-hydroxycytidine. Antivir. Chem. Chemother. 2004;15:15–22. doi: 10.1177/095632020401500102. [DOI] [PubMed] [Google Scholar]
- 36.Ono Y, Saido TC, Sorimachi H. Calpain research for drug discovery: challenges and potential. Nat. Rev. Drug Discov. 2016;15:854–876. doi: 10.1038/nrd.2016.212. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang L, et al. Biochemical characterization of recombinant Enterovirus 71 3C protease with fluorogenic model peptide substrates and development of a biochemical assay. Antimicrob. Agents Chemother. 2015;59:1827–1836. doi: 10.1128/AAC.04698-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai Q, et al. Conformational plasticity of the 2A proteinase from enterovirus 71. J. Virol. 2013;87:7348–7356. doi: 10.1128/JVI.03541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J. Am. Chem. Soc. 2011;133:4274–4284. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marty MT, et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 43.Ma C, Zhang J, Wang J. Pharmacological characterization of the spectrum of antiviral activity and genetic barrier to drug resistance of M2-S31N channel blockers. Mol. Pharmacol. 2016;90:188–198. doi: 10.1124/mol.116.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 45.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Bonuccelli G, et al. Proteasome inhibitor (MG-132) treatment of mdx mice rescues the expression and membrane localization of dystrophin and dystrophin-associated proteins. Am. J. Pathol. 2003;163:1663–1675. doi: 10.1016/S0002-9440(10)63523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mani SK, et al. In vivo administration of calpeptin attenuates calpain activation and cardiomyocyte loss in pressure-overloaded feline myocardium. Am. J. Physiol. Heart Circ. Physiol. 2008;295:H314–326. doi: 10.1152/ajpheart.00085.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng S, Kuang Z, Zhang Y, Xu H, Cheng Q. The protective effects and potential mechanism of Calpain inhibitor Calpeptin against focal cerebral ischemia-reperfusion injury in rats. Mol. Biol. Rep. 2011;38:905–912. doi: 10.1007/s11033-010-0183-2. [DOI] [PubMed] [Google Scholar]
- 51.Akdemir O, et al. Therapeutic efficacy of SJA6017, a calpain inhibitor, in rat spinal cord injury. J. Clin. Neurosci. 2008;15:1130–1136. doi: 10.1016/j.jocn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Li SZ, et al. ALLN hinders HCT116 tumor growth through Bax-dependent apoptosis. Biochem. Biophys. Res. Commun. 2013;437:325–330. doi: 10.1016/j.bbrc.2013.06.088. [DOI] [PubMed] [Google Scholar]
- 53.Delli Pizzi S, et al. Morphological and metabolic changes in the nigro-striatal pathway of synthetic proteasome inhibitor (PSI)-treated rats: a MRI and MRS study. PLoS ONE. 2013;8:e56501. doi: 10.1371/journal.pone.0056501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure for SARS-CoV-2 Mpro has been deposited in the Protein Data Bank with accession number 6WTT.