Figure 1.
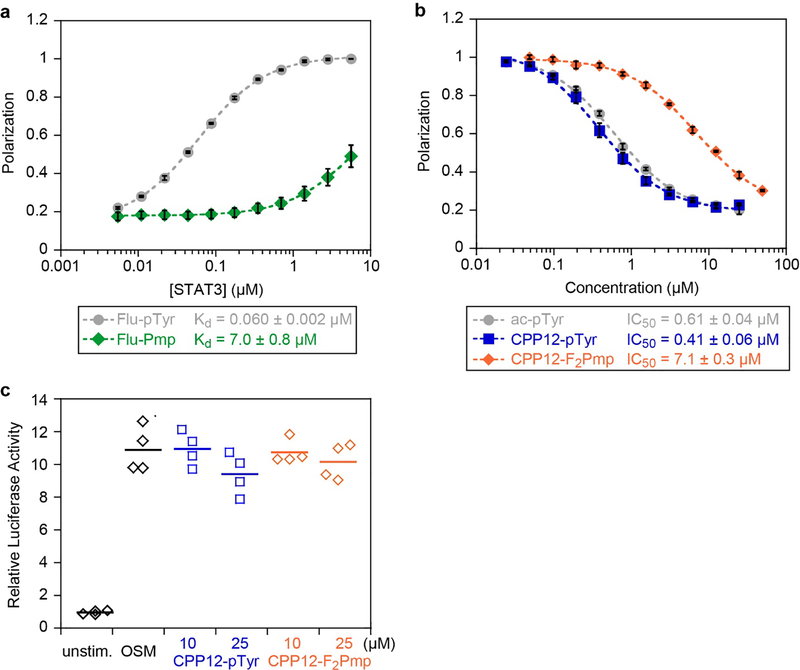
Binding affinities and cellular STAT3 inhibition of selected peptides. a) Fluorescence polarization binding data for selected peptides with recombinant STAT3. Fluorescein-labeled peptides were incubated at 10 nM with serial dilutions of STAT3 at room temperature for 45 min. Data points represent the averages of three biological replicates, each performed with three technical replicates, normalized to the maximum observed polarization (raw data shown in Fig. S1). Error bars show standard error of the mean for the three biological replicates. Kd values are the mean and standard error of the mean for three independent Kd curve fits to the three biological replicates. b) Competition FP for selected peptides with STAT3. flu-pTyr was incubated at 10 nM with 300 nM STAT3 protein and serial dilutions of peptide inhibitors at room temperature for 45 min. Data points represent the averages of three biological replicates, each performed with three technical replicates, normalized to the no-inhibitor control (raw data shown in Figs. S2,S3). Error bars show standard error of the mean for the three biological replicates. IC50 values are the mean and standard error of the mean for three independent curve fits to the three biological replicates. c) STAT3 transcription inhibition. STAT3-luc U3A fibrosarcoma cells were pretreated with 10 or 25 μM of CPP12-pTyr and CPP12-F2Pmp for 24 hours at 37 °C in DMEM supplemented with 10% FBS, followed by 6 hour stimulation with OSM (10 ng/mL). Controls included OSM-treated cells without peptide, and unstimulated cells. This experiment was performed with two biological replicates, each with two technical replicates (all four values displayed).