Figure 1.
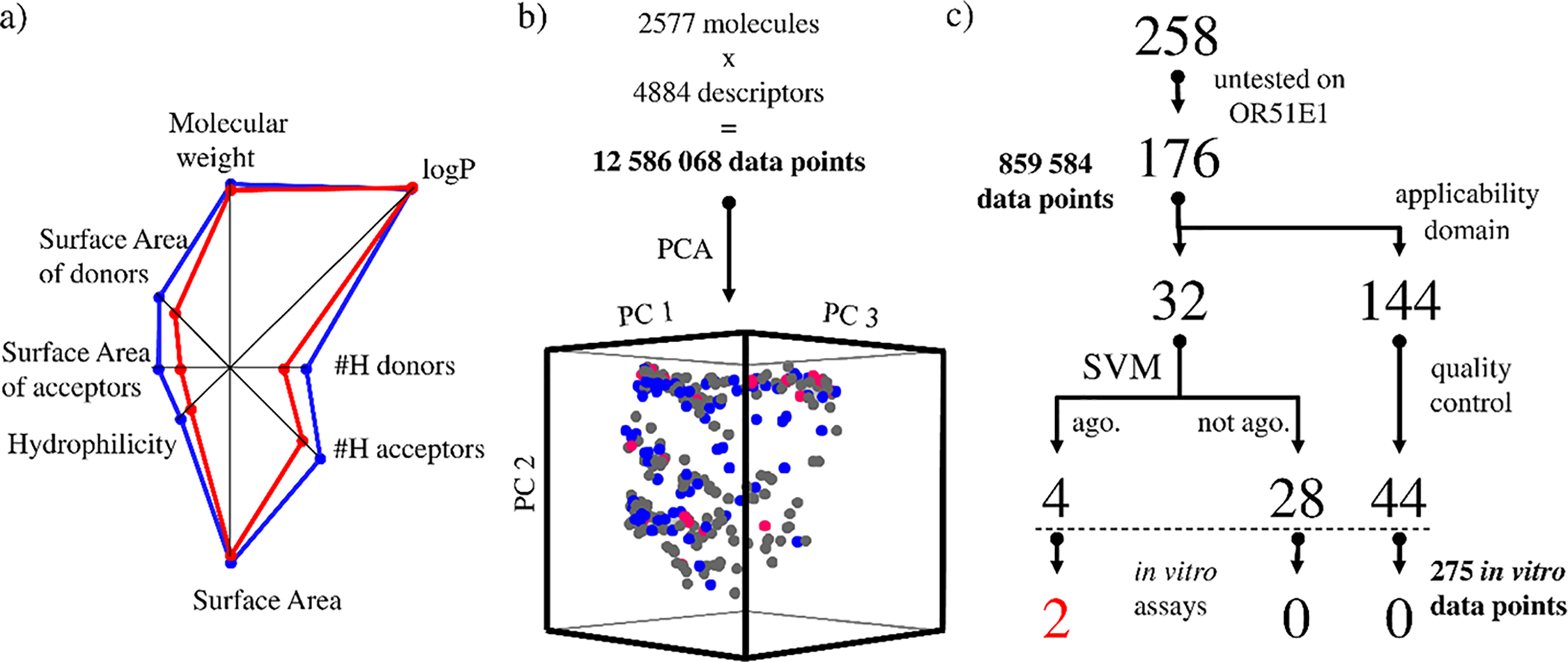
Odorant molecular space and protocol used for OR51E1 virtual screening. (a) Spider plot representing the weight of eight attributes (molecular weight, logP, number of hydrogen-bond donor and acceptors, total surface area, hydrophilicity, and surface areas of acceptor and donor atoms) to define OR51E1 agonists (red) and non-agonists (blue), as identified prior to this study. (b) Projection of our library of 176 original odorants, shown in gray, into the three first principal components of the odorant space computed on the basis of chemical descriptors of 2577 odorant compounds. Known agonists and non-agonists are shown in red and blue, respectively. (c) Of our library of 258 odorants, 176 had been untested on OR51E1, and 32 belonged to the same chemical space (applicability domain) of our model. The virtual screening predicted four agonists, two of which elicited a receptor response in vitro. All predicted non-agonists were tested in vitro for quality-control purposes, as were 44 randomly selected compounds initially excluded from the library; none triggered a receptor response in vitro.
