Figure 3.
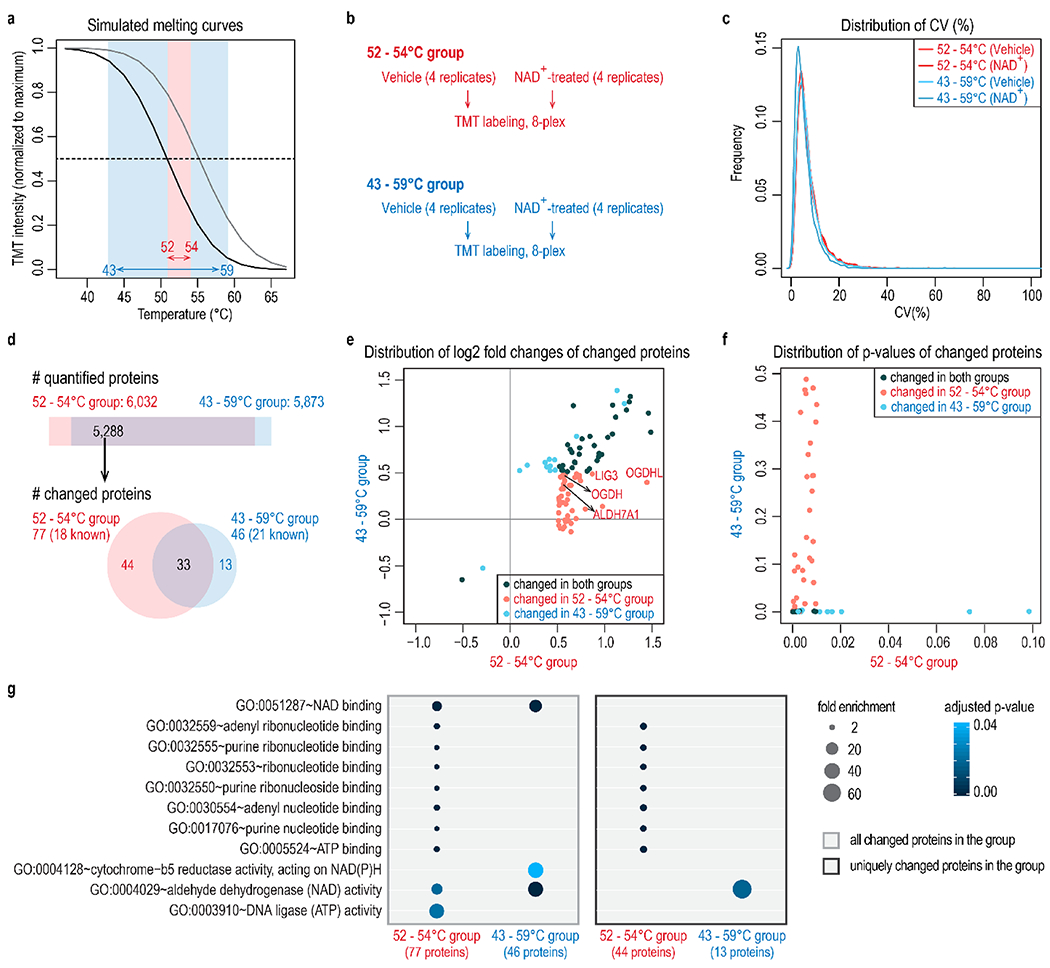
Temperature selection improves the identification of NAD+-binding proteins. (a) Two temperature ranges are compared in this experiment: a broad range of 43 to 59 °C, and a narrow range of 52 to 54 °C. (b) Four replicates of vehicle or NAD+-treated samples were included for each temperature range. Samples were accommodated in two TMT 8-plex experiments. (c) Distribution of coefficient of variation (CV) of four replicates under different conditions shows equivalent data quality. (d) The number of quantified proteins and proteins with altered abundance at different temperature ranges. Both the temperature ranges captured known NAD+-binding proteins. The narrow temperature range of 52–54 °C revealed more targets (77) than the broad temperature range (46). The STITCH database was used to map known NAD+-binding proteins. (e,f) Distribution of log2 fold changes (e) and p-values (f) of changed proteins at either temperature range. The narrow temperature range (52–54 °C) uniquely captured 44 significant target proteins that show small fold changes at a broad temperature range (43–59 °C). Known NAD+-binding proteins uniquely recovered in the 52–54 °C group were highlighted in part e. (g) Gene ontology analysis of proteins with altered abundance in each group. All overlapping quantified proteins were used as the background. p-values were adjusted using the Benjamini-Hochberg method and then filtered at 0.05.
