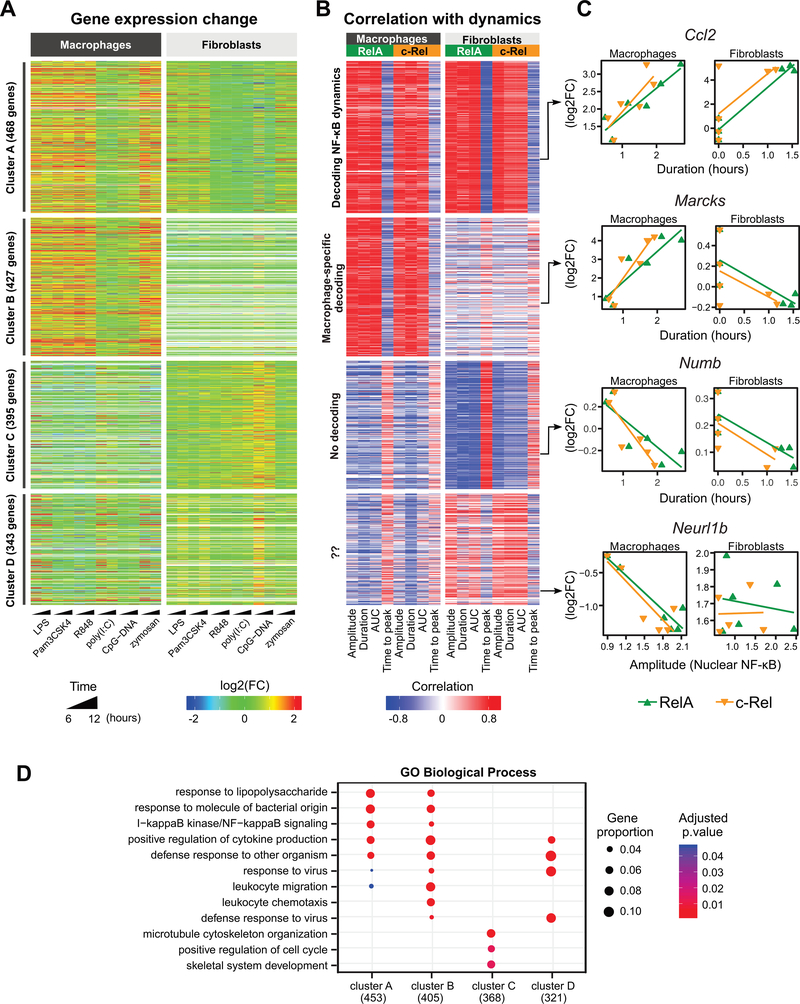
Fig. 4. Dynamic features of NF-κB signaling are linked to gene expression.
(A) Heatmap of the 1633 differentially expressed genes [log2 fold-change (log2FC) ≥ 1 and false discovery rate (FDR) < 0.05 in at least one treatment] obtained from whole-transcriptome analysis of TLR ligand–induced changes in macrophage and fibroblast gene expression. Displayed are the log2FC values of the treated cells relative to unstimulated cells. Each row represents one gene. (B) Pearson correlation values were determined for the log2FC value (mean of the 6- and 12-hour treatments) of each gene with respect to the medians of the single-cell NF-κB dynamics in each cell type and for each NF-κB subunit. Clustering was performed based on the correlation values using partitions around medoids. Values beyond the legend scales were set to extremes. The ordering of the genes is the same as that in (A). (C) Correlations for representative genes derived from each of the four clusters in (A) and (B). Shown are the genes’ expression level changes relative to the median of the single-cell dynamics features for each of the NF-κB subunits in each cell type. The dynamics feature that most strongly correlated with gene expression for each example gene is depicted. (D) Gene Ontology Biological Process (GOBP) enrichment analysis of dynamics-correlated genes in each of the clusters presented in (A) and (B). The three most statistically significant GOBP terms for each cluster were merged and the combined GOBP terms are shown. The number of genes with GOBP annotations are indicated in parentheses. Significant terms are marked as dots. Dot size represents the proportion of cluster genes annotated for each term. P values were adjusted using the default Benjamini-Hochberg method.