Figure 2. Population and phylogenomic analyses of S. cerevisiae, S. kudriavzevii, S. uvarum, S. eubayanus, and their hybrid sub-genomes.
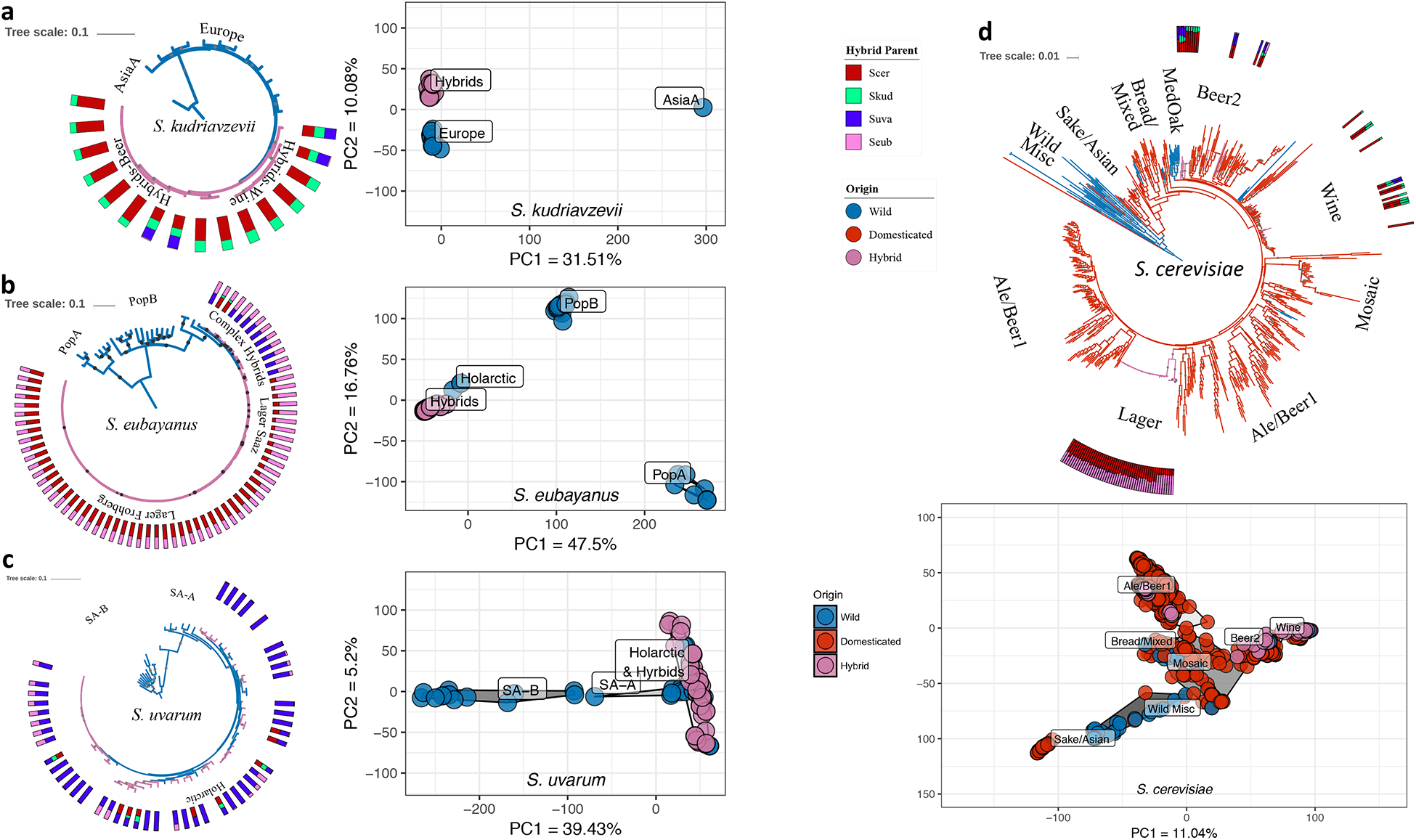
All phylogenies were built with RAxML with pure strains of a species and any hybrids with >50% complete sub-genome for given species. Bootstrap support values >70% are shown as gray dots. Branches are colored by the origin of isolation for each strain. Each hybrid has a stacked bar plot showing the genomic content for each species contributing to their genome; species colors are the same as in Figure 1a. For the Principal Component Analyses (PCA), dot colors represent strains’ origins, and color clouds represent populations or lineages. The axes of all PCAs are scaled to the same range. Phylogenies with strain names, Newick formatted files, and data frames used to build PCAs are available as Figures S2, S4, S6, and S8; Table S2; and Files S1, S3, S5, and S7. (a) Left: Phylogeny of S. kudriavzevii with 30 strains and 38,992 SNPs from across the genome and rooted with an Asia B strain, IFO1803 (removed for clarity). Right: Principal component projection for PC1 and PC2, excluding Asia B. (b) Phylogeny of S. eubayanus with 92 strains and 18,878 SNPs from across the genome, rooted with Population A (PopA). Right: Principal component projection of PC1 and PC2. (c) Phylogeny for S. uvarum with 82 strains and 18,652 SNPs from across the genome, rooted with the Australasian lineage (removed for clarity). Right: Principal component projection for PC1 and PC2, excluding the Australasian lineage. (d) Top: Phylogeny for S. cerevisiae with 612 phased (for strains with >20K heterozygous sites) or unphased haplotypes and 21,222 SNPs from across the genome, rooted with the Taiwanese strain EN14S01 (removed for clarity). Previously identified wild lineages from West Africa, Malaysia, North America, Japan, and the Philippines are included in the Wild Misc group11,74. The other lineages are named in a similar manner to previous studies on ale-brewing and Mediterranean Oak (MedOak) strains8,9,71. Bottom: Principal component projection for PC1 and PC2 (including EN14S01, which groups with Sake/Asian).
