Figure 3. Mitochondrial genome inheritance in interspecies hybrids.
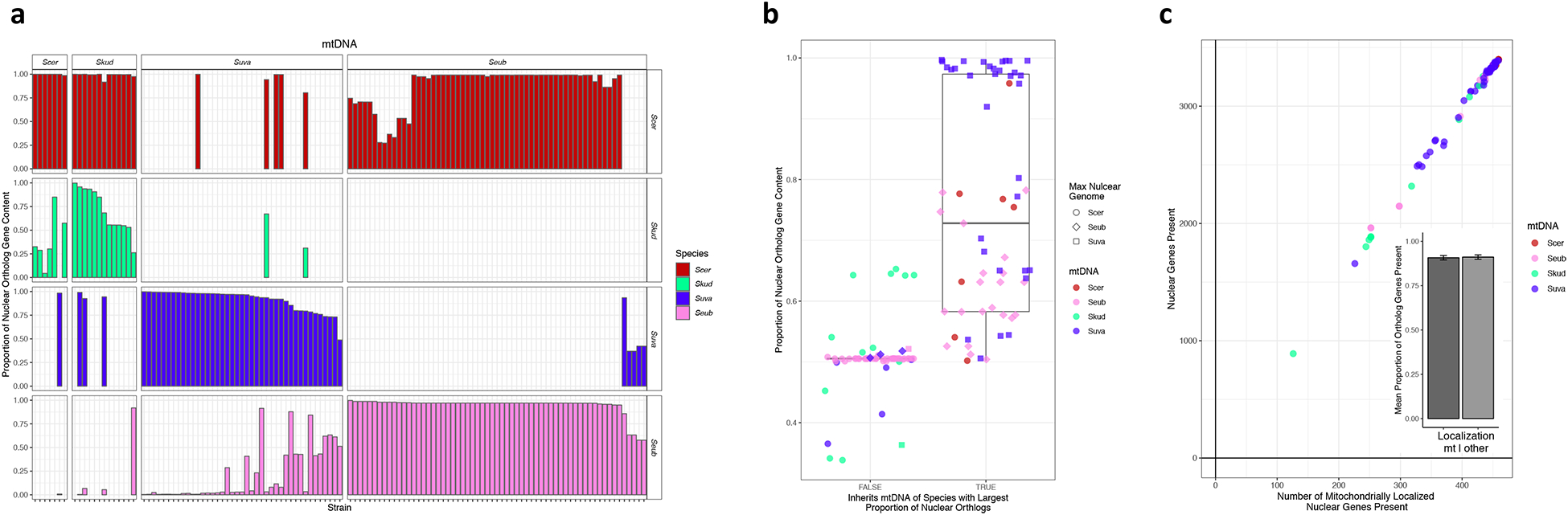
(a) The bar plots show proportion of 1:1:1:1 ortholog content for each sub-genome for each hybrid grouped by the mitochondrial genome (mtDNA) parent, which are labeled across the top. Colors represent different parent species and are that same as in of Figure 1a. (b) Analysis of concordance between which mtDNA was inherited and which parent contributed the most complete set of orthologous genes. “True” includes hybrids that inherited the most nuclear gene content from the same species as the mtDNA. “False” includes hybrids with mtDNA that did not match the species that contributed the most nuclear gene content. Colors represent the mtDNA parent, and shapes represent the largest nuclear genome contributor. The middle of the box plot corresponds to the median, the upper and lower limits are the 75th and 25th percentiles respectively, and the whiskers extend to the largest or smallest value no greater than 1.5 × the differences between the 75th and 25th percentiles. There was a significant correlation between the mtDNA parent and the largest nuclear genomic contributor (logistic regression p=3.58E-8, AIC= 118.21). Notably, the S. eubayanus × S. uvarum hybrids, which have often undergone many backcrossing events, follow this trend and are both cryotolerant species. (c) Linear relationship of the number of 1:1:1:1 orthologs versus the number of nuclear-encoded, mitochondrially localized genes present in the sub-genome that matches the mtDNA (linear regression p=2.0E-16, AIC= 1151.5). The inset shows the mean proportion of mitochondrially localized versus all other nuclear genes present in the sub-genome that matches the mitochondrial parent (p = 0.8612, odds ratio = 0.9653).
