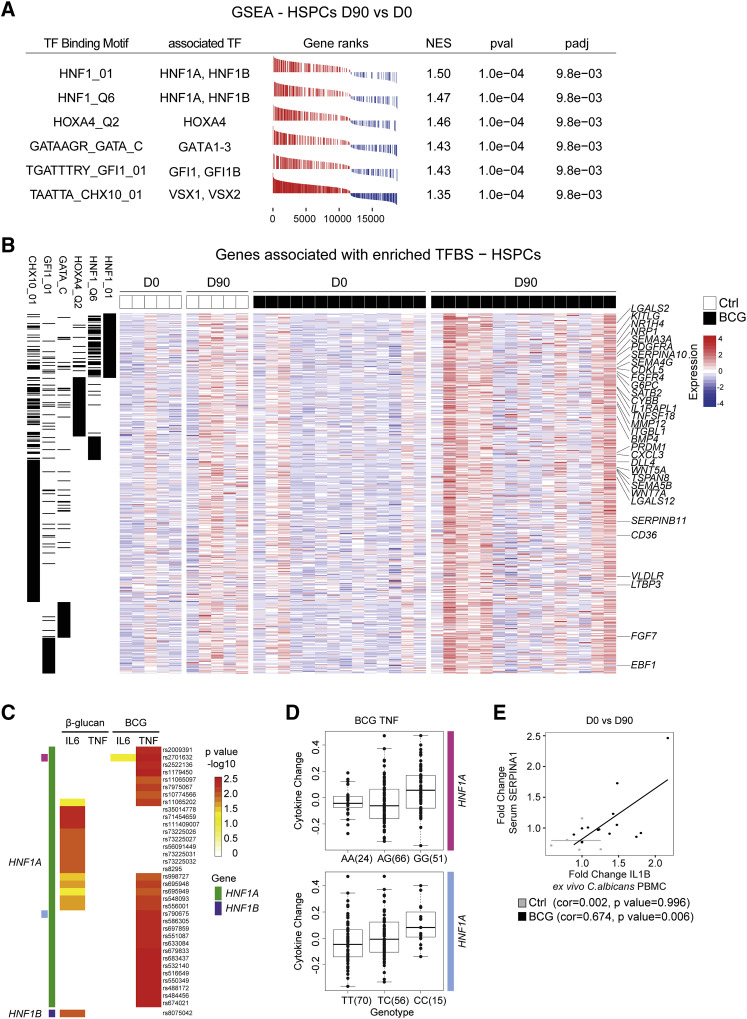
Figure 5.
HNF1 TFs Represent Regulatory Factors in Trained Immunity
(A) GSEA of enriched signatures associated with distinct TF-binding motifs in genes ranked according to expression fold change in HSPCs at D0 versus D90. TFs associated with the binding motifs are listed. NES, normalized enrichment score; pval, p value; padj, adjusted p value.
(B) Expression heatmap of all leading-edge genes derived from TF-binding site gene sets as found in (A). Black lines in the first six columns indicate association of genes to the respective TF signature gene sets listed in the column header.
(C) Ex vivo training of PBMCs with β-glucan or BCG and IL6/TNF cytokine measurements after LPS challenge. SNPs ± 250 kb within the genomic locus of HNF1A (green) or HNF1B (purple) and variants thereof significantly leading to a change in cytokine production for any of the conditions are listed. Threshold for inclusion, p < 0.01. Top two expression quantitative trait loci (eQTLs) are annotated as pink and bright blue.
(D) Detailed TNF cytokine level after BCG training and LPS challenge for the top two eQTLs (annotated in C as pink and bright blue). Number of individuals displaying distinct SNP variants are shown in brackets. Analysis in (C) and (D) is based on 141 individuals in total.
(E) Fold changes of IL1B after ex vivo C. albicans restimulation and serum SERPINA1 (D0 versus D90) correlated in the same individuals. Data points from BCG-vaccinated or Ctrl-treated individuals are shown in black or gray, respectively.