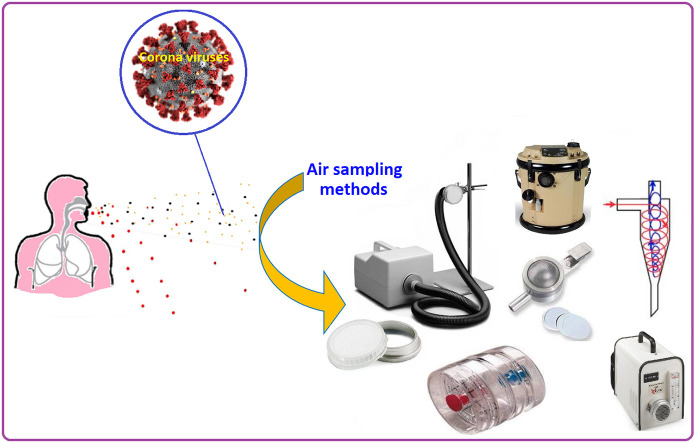
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a strain of coronaviruses that causes coronavirus disease 2019 (COVID-19). In these days, the spread of the SARS-CoV-2 virus through the air has become a controversial topic among scientists. Various organizations provide standard methods for monitoring biological agents in the air. Nevertheless, there has been no standard recommended method for sampling and determination of viruses in air. This manuscript aimed at reviewing published papers for sampling and detection of corona viruses, especially SARS-Cov-2 as a global health concern. It was found that SARS-Cov 2 was present in some air samples that were collected from patient's rooms in hospitals. This result warrants its airborne transmission potential. However, due to the fact that in the most reviewed studies, sampling was performed in the patient's room, it seems difficult to discriminate whether it is airborne or is transmitted through respiratory droplets. Moreover, some other disrupting factors such as patient distance from the sampler, using protective or oxygen masks by patients, patient activities, coughing and sneezing during sampling time, air movement, air conditioning, sampler type, sampling conditions, storage and transferring conditions, can affect the results. About the sampling methods, most of the used samplers such as PTFE filters, gelatin filers and cyclones showed suitable performance for trapping SARS-Co and MERS-Cov viruses followed by PCR analysis.
Keywords: SARS-Cov-2, Virus, Air, Sampling
Graphical abstract
1. Introduction
Microorganisms can disperse into the air through dust and water drops. Some bacteria are mainly considered as airborne microorganisms. However, other microorganisms such as algae, protozoa, yeasts, molds, and viruses have been isolated from air samples (Sandle, 2016). Most of the previous studies have shown that outbreaks can be controlled by increasing hand hygiene and disinfecting the environment. However, transmission of infection by the air has been less well investigated. Mycobacterium tuberculosis is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis. This bacterium can be easily transmitted through air, and hence is considered a source of outbreak in hospitals. Many previous studies have focused on the airborne bacteria and few studies have evaluated the presence of viruses with pathogenic effects in the air (Beggs, 2003; Leung and Chan, 2006). It has been reported that hospital-acquired respiratory viral infections are a major cause of morbidity and mortality among hospitalized patients (Chow and Mermel, 2017), which highlights the importance of regular sampling and monitoring of virus load in indoor air and surfaces of hospitals. Coronaviruses are a group of related RNA viruses with a large genome. Among severe acute respiratory syndrome coronaviruses (SARS-Cov viruses), which cannot be regarded as a seasonal virus, there are four HCoVs, originally divided into two serogroups (the third serogroup does not contain any human viruses), that contribute to seasonal outbreak (Campanacci et al., 2003; Li et al., 2005). Severe acute respiratory syndrome (SARS) is a viral respiratory illness that was first identified in 2003. SASR virus can easily spread through reparatory droplets like flu viruses (Drosten et al., 2003). Until now, few studies have focused on the prevalence of respiratory viral infections as a hospital-acquired infection. In this regard, a previous study on the prevalence of acute respiratory infections among hospitalized patients showed that viruses are associated with a significant percentage of acute respiratory infections (Kouni et al., 2013; Shi et al., 2015). Moreover, a similar study reported that of five children admitted to intensive care unit, one child had acquired a viral respiratory infection. They also reported that the viral respiratory infection could 6-fold increase the mortality rate in comparison with those had acquired viral a respiratory infections from community (Spaeder and Fackler, 2011). It has been also reported that 24% of pneumonias in hospitalized patients were caused by viruses in nonventilated hospital. Due to the lack of specific treatment for most of viruses, it is essential to monitor the level of virus in indoor air of hospitals, especially in different wards with high patient density. On the other hand, the determination of the personal exposure of medical stuff to viruses as a type of occupational exposure is very important, which not enough attention was given to regular monitoring of these pathogens in hospital as an important occupational environment. Occupational health organizations have introduced and recommended a variety of methods for monitoring bioaerosols in the air of occupational environments (Lindsley et al., 2017). The National Institute for Occupational Hygiene (NIOSH) has introduced Anderson bioaerosol sampler for sampling and identification of culturable microorganisms and assessment of possible proliferation and dissemination of bacteria or fungi in workplaces (NIOSH, 1998). In the NIOSH recommended method, Andersen 2-stage cascade impactor has been introduced for sampling of fungi and mesophilic bacteria, and Andersen N-6 single-stage sampler for thermophilic actinomycetes (NIOSH, 1998). The American Industrial Hygiene Association (AIHA) has also recommended a field guide for the monitoring biological agents in environments (Dillon et al., 2005). The AIHA has recommended that microbial air sampling might be conducted during the outbreak of respiratory infection disease. It has been also reported that microbial air sampling had a useful role in the etiologic agent of many bacterial respiratory infections (Dillon et al., 2005). So far, no method has been recommended by the related organizations for the sampling and determination of viruses in the air of workplaces. While as mentioned above, viruses have a main role in acute respiratory infections in hospital as an important workplace. In air sampling of bacteria, a suitable sampling medium for the target microorganism is necessary to support the growth of trapped microorganisms (Chinn and Sehulster, 2003). In contrast, viruses are not able to grow in general sampling media, because of their structure that consists of a DNA or RNA genomes encased in a fatty liquid membrane and need a host cell to reproduce and cause infection (J Alsaadi and Jones, 2019). The use of sampling media is still a challenge to sample viruses from air. Therefore, it the present study, we tried to review recent methods for sampling and determination of viruses in air by focusing on the corona viruses, especially SARS-Cov-2 as a global health concern. Moreover, in this paper, the effect of various parameters on the sampling of viruses such as sampling tools, sampling time, flow rate, type of culture medium and requirements for storage and transfer are discussed. Due to the lake of standardized method for identification of SARS-Cov-2 virus, in the present study, we tried to review the successful methods that have been used for sampling and detection of SARS like viruses in the air.
2. Sampling conditions
No single sampling procedure is appropriate for sampling of all indoor bioaerosols. There are a lot of bioaerosols that their behavior is different in the air. Bioaerosol behavior is associated with particle size, and hence selecting an appropriate sampling procedure is based on the particle size and bioaerosol type. For sampling a specific type of viruses in the air, different types of sampling device, sample pump, sampling volume, sampling time, culture medium and incubation conditions are used. Each of the sampling methods can provide different performances for the sampling and detection of various types of viruses. Selecting a sampling procedure should be done based on the prevention of damage to the microbial agents. In these cases, it is necessary to minimize microbial stress during the sampling period (such as desiccation effects and strong impaction at long sampling time). In air sampling of RNA viruses, maintaining the viability of the virus is not mandatory, but maintaining its nucleic acid integrity is essential. Because the nucleic acid can rapidly be degraded during sampling (Verreault et al., 2008). In addition to sampling procedure, sampling conditions play a key role in the accurate sampling and detection of the target virus. In the air sampling of SARS-like viruses, various factors such as ventilation, air movement, distance from the patient bed, occupancy and patient activities during the sampling, relative humidity, temperature, number of patients in the sampling room, sampling flow rate, sampling time, and sampling medium, can affect the results (Cox et al., 2020). In this regard, for determining a suitable and standard protocol for the sampling and detection of SARS-Cov-2, the type of sampler and the sampling environmental conditions during sampling should be considered simultaneously. These sampling conditions are only controllable in laboratories to provide a constant condition for investigating the influence of each parameter on the sampler performance. In sampling of SARS-Cov-2 in indoor of hospital, additional factors must be also considered such as noise level produced by sampling pumps, air movement, wearing protective and oxygen masks by patients that prevent releasing their respiratory droplets in sampling site, time needed for air sampling, minimal interruption to the medical care service, etc. Therefore, it is difficult to define a sampling condition and identify a specific and suitable protocol for determining SARS-Cov-2 in indoor air of hospital. Moreover, inconsistent results have been reported in the use of various sampling procedures for the detection of SARS-Cov-2 in air.
2.1. Sampling procedure
In the first step in the sampling of viruses in air, it is essential to know that the viruses are not transmitted independently of particles in the air. The particles containing viruses consist of a mixture of different compounds, such as organic and inorganic matter, proteins, salt and viruses (Verreault et al., 2008). Therefore, the size of viruses is different from the particle containing viruses. Hence, sampler should be selected based on the detection of viral particle size, so that filters with a pore size larger than viruses can be used for sampling of airborne viruses in air. Most of the previous studies have classified sampling methods of viruses into two categories including dry and wet sampling methods. In the dry sampling method, a filter membrane is commonly used in two closed and open face filter cassettes. Filter cassettes are generally used in two types including 2-Stage and 3-Stage cassettes. There are inlet and outlet ports in the cassettes for entering and exiting air after passing through the filter. The 2-Stage cassette can only be used for closed faced sampling, while the 3-Stage cassette is used for the both closed and open faced sampling. Some studies have used polytetrafluoroethylene (PTFE) membrane filters for sampling SARS like viruses (Booth et al., 2005). However, other types of filters like tightly packed cotton and cellulose filters have been also applied for sampling airborne viruses (e.g. Poxviridae family) in air (Meiklejohn et al., 1961). In a previous study, the efficiency of three different filters including PTFE, polycarbonate (PC) and gelatin filters was evaluated for sampling bacterial and viruses in air, and the concentration of viral particle size was measured to determine the physical collection efficiency of the filters. In this case, the PTFE and gelatin filters showed the highest performance for the sampling of the viral particle size, while the PC filter offered the lowest performance for the collection of the viral particle size. Therefore, filter material play a key role in the collection efficacy of filters for sampling viral particle size (Burton et al., 2007). It should be noted that gelatin filters are used in MD-8 air scanner, which is portable and modern air sampling instrument for the collection of airborne microorganisms or viruses. Moreover, the NIOSH in a recent report has reviewed various filters for sampling airborne pathogens and recommended the use of PTFE for immunological assays and polymerase chain reaction (PCR). Due to the structure of PTFE filters, they do not interfere with biochemical tests and the target viruses, and hence can be easily eluted from the membrane (Lindsley et al., 2017). Despite the widespread use of filters in air sampling of coronavirus-like particles, some restrictions have been reported for their application in air sampling of viruses such as being very destructive for the virus. However, filters offer a high physical collection efficiency for virus-containing particles (Pan et al., 2019). It should be noted that applying high volume requires high power and may produce undesired noise which in some cases is not applicable such as sampling in intensive care units (ICUs). High volume sampling requires high volume sample pumps that are not easy portable, and in longer sampling time the desiccation effects can cause damage to the target viruses. Also, applying high sampling volume with gelatin filter may dissolve the gelatin into water, and thereby affect the sampler performance (Pan et al., 2019). In addition, sampling with the gelatin filter at low relative humidity can cause desiccation of viruses, while high relative humidity can lead to dissolution of gelatin filters. These points should be taken into account during sampling with gelatin filer as a commonly used method for air sampling of SARS-Cov-2 viruses (Fabian et al., 2009).
Biosamper is a bioaerosol collection device that can collect bioaerosols in a liquid for sample times up to 8 h. The Biosampler have used in most of recent papers as a preferred sampling device for air sampling of SARS like viruses (Kim et al., 2007). The biosampler consists of an inlet, outlet, collection vessel in the bottom section and three nozzles (sonic orifices). The Biosampler nozzle section contains three tangential 0.630-mm nozzles that act as sonic orifices. The air flows through the nozzles and remains constant at approximately 12.5 L/min when used with a pump that maintains a pressure drop of 0.5 atm (15 in Hg) or more across the sampler at normal atmospheric conditions (sonic flow). The Biosampler operating instruction has recommended the use of an environmental vacuum sample pump (high flow rate pump in the range 10–60 L/min) for collecting air with the Biosampler. Therefore, this sampling tool is not suitable for personal sampling because of using the environmental vacuum pump that is connected to the impinger/Biosampler. Moreover, impingers might not be applicable for sampling of the virus at low viral load because of its limited flow rate capacity. Various collection liquids, such as sterile distilled water, physiological saline, phosphate buffered saline (PBS), nutrient broth, and peptone water, have been recommended for the collection of target organisms in the Biosampler based on the sampling time. Some studies have also used liquid impingers like Biosampler for air sampling viruses. In a review study, the performances of filter membrane and liquid or glass impingers were compared and it was reported that the impinger like samplers were the most efficient sampler, while filters were very destructive for bacteriophage as an indicator of other viruses in the air (Verreault et al., 2008). However, it should be noted that the PCR analysis can detect non-infectious viruses that have been destructed during sampling with filter based methods. But, this point can be considered for other analyses that require infectious viruses for further investigations. In a recent study, the impinger technique was used for the sampling of SARS-Cov-2 in the indoor hospital environment. In the mentioned study, an average flow-sample pump was used instead of a high volume vacuum sample pump and the impinger was filled with 20 mL of Dulbecco's Modified Eagle's Me-dium (DMEM), streptomycin (100 μg/mL), penicillin (100 U/mL), and 1% anti-foam reagent (isoamyl alcohol). Isoamyl alcohol has been used as an anti-foaming reagent to prevent impinger liquid (culture medium) from being drawn into the sample pump (Faridi et al., 2020). However, such problem has not been addressed in the previous studies and a mixture of culture medium and antibiotics have been used without adding anti-foaming reagent (Kim et al., 2016). It has been reported that impinger like sampler are the most commonly used samplers for sampling viruses in air. Because, the liquid impinger helps to maintain the integrity and viability of the virus and can be directly analyzed without the need for preparation and extraction of the target virus from a filter surface and or medium (Pan et al., 2019). However, they might not be appropriate for sampling low level of viruses in the air due to their limited flow rate capacity. Foam production caused by the presence of viral culture medium in impinger is another limitation in its application for air sampling of viruses, which has been addressed in Faridi et al. (2020) study. According to the Biosampler operating instructions, sterile distilled water, physiological saline, PBS, nutrient broth, or peptone water can be used in Biosampler (as an impinger-like sampler) for sampling bioaerosols even without trap and anti-foaming agent.
Cyclone sampler has been severally used for sampling of SARS like viruses. The cyclones were used for 8 h and high flow rate for sampling such viruses. Moreover, NIOSH method 0600 has recommended the use of cyclone prior filter for removing the non-respirable particles from the target bioaerosols (Lindsley et al., 2017). In a developed method, the NIOSH has recommended the use of two consecutive cyclones for accurate separation of non-respirable particles prior to the main filter. The NIOSH cyclone has been applied for airborne viruses in hospital environments (Blachere et al., 2011). Some studies have reported that the cyclone methods are very destructive for collecting viruses. However, as mentioned above, PCR analysis can detect non-infectious viruses, and therefore there is no need to trap infectious form of the airborne viruses. A recent study has used cyclone along with filter cassette containing a 37-mm diameter PTFE filter with 3 μm pore sizes for sampling and determination of SARS-Cov-2 viruses in the indoor air of hospital. In this case, the cyclones were used prior to the filter cassette containing PTEF filter membrane for the distribution of the collected particles in three size fractions including >4 μm, 1–4 μm and <1 μm (Chia et al., 2020). Physical damage resulting from the actions of cyclones during sampling is considered a main limitation in their application for virus-containing particles.
MD-8 air scan sampling device has been also applied for sampling of MERS-coronavirus (MERS-CoV) and SARS-Cov viruses in indoor air of hospitals (Kim et al., 2016). This sampler works based on the filtration principle. In this method, a determined volume of sampling air is drawn through a gelatin filter and after sampling the gelatin filter is dissolved in viral transport medium to retain the trapped viruses for virological or molecular biology techniques. The gelatin filter with a nominal pore size of 3 μm, 80 mm in diameter and soluble in water can be used in the MD-8 air scan sampler (Kim et al., 2016). Until now, various studies have used the MD-8 air scan sampler for sampling of MERS-CoV viruses in air. It has been reported that the MD-8 airscan sampler offered a 100% collection efficiency for RNA virus (aerosolized infectious bursal disease virus: IBDV) (Zhao et al., 2014). Moreover, in a recent study in Singapore, side by side sampling with the MD-8 airscan and PTFE filter placed in a cassette was performed in airborne infection isolation rooms (with 12 air exchanges per hour) for sampling and detection of SARS-Cov-2 viruses followed by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) analysis, and they reported that all air samples were negative (Ong et al., 2020). In Agranovski et al. study, a novel method based on air sampling with HEPA filters was introduced for sampling and determination of SARS like viruses in indoor air of hospital. For this reason, a HEPA filters was installed in the pipeline connecting sampler and vacuum pump. Moreover, Booth et al. (2005) used a high-resolution slit-sampler system designed by Defense Research and Development Canada (DRDC). In this method, a 0.15 × 48 mm slit with a rate of 30 L/min was impinged onto a 150-mm petri dish with a 12% gelatin base containing viral collection medium (sterile phosphate buffer with 7.5% bovine serum albumin, 10,0000 U/mL penicillin G, 10,000 mg/mL streptomycin sulfate, and 25 mg/mL amphotericin B). It should be noted that in the mentioned study, side by side sampling was performed using the proposed high-resolution slit-sampler system as wet sampler and PTFE filter placed in a cassette as dry sampler. According to the reviewed papers, gelatin filter have been commonly used in MD-8 air scan and showed an appropriate performance for sampling corona viruses. In a recent study, the gelatin filter was successfully used with simple styrene cassette at a flow rate of 5 L/min (Liu et al., 2020).
2.2. Sampling time and flow rate
A wide range of sampling times has been recommended for sampling bioaerosols in air. In the NIOSH recommended method for sampling of bacteria, fungi and thermophilic actinomycetes, there are no specific recommended limits of sampling time and flow rate for bioaerosol sampling (indoor air) (Lindsley et al., 2017). However, the Andersen sampler works with a constant flow rate (28.3 L/min) and default timer for sampling a determined volumes of air (10 min). To the best of our knowledge, no study has used Anderson sampler for sampling of SARS like viruses in air. In a previous study, it has been reported that the Andersen impactor did not obviously inactivate virus during sampling and then can be used for sampling airborne viruses (Zhao et al., 2014). Faridi et al. (2020) have used an impinger attached to an average flow rate of 1 L/min for 1 h, and did not find any SARS-Cov-2 virus in the indoor air samples. Impinger sampler might not be an ideal solution for sampling the virus with low viral load, because it has a low flow rate and is not applicable at very high flow rates. In a previous study in the detection of airborne SARS coronavirus, a high-resolution slit-sampler system designed by Defense Research and Development Canada was applied with a flow rate of 30 L/min for 18 min, which reported that 1 of 10 air samples were positive for SARS-Cov. It seems that due to the low concentrations of SARS like viruses in air, a relative high volume of air should be sampled for PCR analyses. It should be noted that in the mentioned study, the hospitalized patients was requested to not wear a mask and had normal activities (Booth et al., 2005). Therefore, these results confirmed the presence of SARS corona virus in the air of the patient's rooms. Moreover, in Kim et al. (2016) study, an MD-8 airscan sampling device was applied for detecting virus in indoor air of hospital. In the mentioned study, the sampler was applied with a flow rate of 50 L/min for 20 min, and it was found that 4 of 7 air samples were PCR positive for MERS-Cov virus (Kim et al., 2016). In another study, gelatin filter was utilized in cassette for sampling of SARS-Cov-2 in indoor air of hospital with a flow rate of 5 L/min for 1 h, and high load of this virus was identified in the toilet areas used by the patients and low viral load observed in in isolation wards (Liu et al., 2020). Ong et al. (2020) reported negative results in terms of the presence of SARS-Cov-2 virus in air. They have used PTFE filter cassette with a flow rate of 5 L/min for 4 h and MD-8 microbiological sampler with 6 m3/h (100 L/min) for 15 min (Ong et al., 2020). In the method introduced by Agranovski et al. (2004) the sampler was applied at 4 L min−1 for 4 h. In Dong Guo et al. study, a SASS 2300 Wetted Wall Cyclone Sampler was applied with a flow rate of 300 L/min for of 30 min, which some air samples were PCR positive for SARS-Cov-2 (Guo et al., 2020). This finding can be attributed to the high sampling flow rate compared with the previous studies. According to Table 1 , in a total sampling volume of 9000 L, MERS-Cov virus has been identified by Azhar et al. (2014) which indicated that a high sampling volume increases the chances of trapping virus in the indoor air. Cyclone samplers have also been used at high flow rates, ranging from 700 to 1000 L/min (5- to 30-min samples), to recover airborne viruses. In this regard, Guo et al. (2020) identified SARS-Cov-2 in indoor air of hospital using cyclone sampler at high sampling flow rate and time. In Booth et al. (2005) study, the high-resolution slit-sampler system was used for 18 min with a flow rate of 30 L/min. In a study that was conducted on the detection of MERS-Cov virus in indoor air of a camel barn owned by an infected patient, some air samples were PCR positive when one of the nine camels was present in the barn (Azhar et al., 2014). Given the reviewed methods, it can be concluded that the SARS and MERS corona viruses were only detectable in close proximity to the patients and a relative high volume of air containing corona viruses should be sampled for PCR analyses.
Table 1.
Methods and conditions of corona sampling of viruses in air.
| Study | Sampler | Target virus | Sampling time | Sampling flow rate (sampling volume) | Transport and storage conditions | PCR results | |
|---|---|---|---|---|---|---|---|
| 1 | (Faridi et al., 2020) | Impinger attached to a personal sample pump with average flow | SARS-Cov-2 | 1 h | 1 L/min (60 L) | 20 mL of DMEM, 100 μg/mL streptomycin, 100 U/mL penicillin and 1% isoamyl alcohol | Negative |
| 2 | (Kim et al., 2016) | MD-8 airscan sampler | MERS-Cov | 20 min | 50 L/min (1000 L) | in 30 mL viral transport medium (sterile phosphate buffer with 10% fetal calf serum, 10,000 U/mL penicillin, 10 mg streptomycin, 25 μg amphotericin B) and stored at −80 °C until analyzed. | 4 of 7 air samples were positive |
| 3 | (Ong et al., 2020) | MD-8 airscan sampler | SARS-Cov-2 | 15 min | 6 m3/h (1500 L) | – | Negative |
| 4 | (Ong et al., 2020) | 37-mm filter cassettes and 0.3-μm polytetrafluoroethylene filters | SARS-Cov-2 | 4 h | 6 m3/h (400 L) | – | Negative |
| 5 | (Agranovski et al., 2004) | HEPA filters installed in the pipeline connecting sampler and vacuum pump | SARS coronaviruses | 4 h | 4 L/min (960 L) | Vero cell culture produced from the kidney of African green monkey was used for cultivation of SRC VB “Vector”. Inoculated cell cultures were maintained with RPMI 1640 supplemented with 1% fetal bovine serum for TEM analysis, and a mixture containing 2% volumetric of inactivated bovine serum, 100 U ml−1 of penicillin and 100 μg ml−1 of streptomycin the maintenance fluid for collecting virus | Positive |
| 6 | (Guo et al., 2020) | SASS 2300 Wetted Wall Cyclone Sampler | SARS-Cov-2 | 30 min | 300 L/min (9000 L) | – | Positive |
| 7 | (Chia et al., 2020) | Cyclone Bioaerosol Sampler | SARS-CoV-2 | 4 h | 3.5 L/min (840 L) | At 4 °C in hospital prior to transfer to laboratory, and then stored at −80 °C unless directly analyzed | Positive |
| 8 | (Booth et al., 2005) | PTFE membrane filter with a pore size of 0.3 μm in a closed-face, 3-piece disposable plastic cassette attached to a personal sample pump | SARS coronavirus (SARS-Cov) | 10.5–13 h | 2 L/min (1260–1560 L) | Shipping samples with ice packs and refrigerating at the laboratory | Positive |
| 9 | (Booth et al., 2005) | High-resolution slit-sampler system designed by Defence Research and Development Canada (DRDC) (impinger like sampler) | SARS coronavirus (SARS-Cov) | 18 min | 30 L/min (540 L) | Shipping samples with ice packs and refrigerating at the laboratory | Negative |
| 10 | (Azhar et al., 2014) | MD-8 airscan sampling device (Sartorius) and sterile gelatin filters | MERS-Cov | 20 min | 50 L/min (1000 L) | Gelatin filters were dissolved in 5 ml viral transport medium (VTM) and stored at −80 °C until analyzed. | Positive |
| 11 | (Liu et al., 2020) | Sterilized gelatin filters with pore size 3 μm placed in styrene filter cassette | SARS-Cov-2 | 1 h | 5 L/min (300 L) | Filters were preloaded inside the samplers in Class 100 sterilized room and sealed with Teflon tapes | Positive |
2.3. Sampling culture medium
Sampling culture media in the air sampling of corona viruses are used for surviving them for further analyses. Unlike most bacteria that can be grown in artificial media such as nutrient broths and agar plates, viruses cannot be grown on an artificial culture and need a host cell to reproduce and cause infection. In this regard, the constituents of suitable viral transport media are designed to provide an isotonic solution containing protective protein, antibiotics to control microbial contamination, and one or more buffers to control the pH. Liquid transport media are used primarily for transporting swabs or materials released into the medium from a collection swab. Liquid media may be added to other specimens when inactivation of the viral agent is likely and when the resultant dilution is acceptable (Johnson, 1990).
In air sampling of corona viruses, especially SARS-Cov and SARS-Cov-2, various types of viral transport media have been used. The type of the transport medium depends on the sampling method. For example, in impinger like samplers, a mixture of DMEM and antibiotic agents are commonly used as the liquid impinger to collect and survive the target viruses. As mentioned above, DMEM is the most broadly suitable medium for many adherent cell phenotypes, which can trap viruses during air sampling with impinger like samplers and then survive them. Some previous studies have used an anti-foaming agent such as isomyl alcohol to prevent foam formation and its leakage into the sample pump. In this regard, a recent study has used a mixture of DMEM, streptomycin, penicillin, and isoamyl alcohol (anti-foaming reagent) to sample SARS-Cov-2 in the indoor air of hospital (Faridi et al., 2020). In the method introduced by Agranovski et al., Vero cell culture produced from the kidney of African green monkey was used for cultivation of SRC VB “Vector” and then the inoculated cell cultures were maintained with RPMI 1640 supplemented with 1% fetal bovine serum. In the mentioned study, Transmission Electron Microscopy (TEM) analysis was used to investigate the morphological characteristics (Agranovski et al., 2004). In this method, the target virus must be cultured in a suitable medium to obtain higher concentration of the virus. Because, as previously mentioned, viruses cannot be grown on an artificial culture and need a host cell to reproduce and cause infection (Agranovski et al., 2004). In another study, impinger liquid containing 20 mL of DMEM, 100 μg/mL streptomycin, 100 U/mL penicillin and 1% isoamyl alcohol was used as sampling culture medium (Faridi et al., 2020). Moreover, in Kim et al. study, 30 mL of a mixture containing sterile phosphate buffer with 10% fetal calf serum, 10,000 U/mL penicillin, 10 mg streptomycin, and 25 μg amphotericin B, was used as viral transport medium (Kim et al., 2016). In Agranovski et al. study, two different liquids were applied for preparation of viral transport media; a mixture containing 2% volumetric of inactivated bovine serum, 100 U mL−1 of penicillin and 100 μg mL−1 of streptomycin the maintenance fluid prepared from Hank's solution containing was applied to provide a suitable conditions to reduce damage the virus. In order to avoid foam formation, Antifoam A as an anti-foaming agent was added to the viral transport media for trapping the SARS-Cov virus (Agranovski et al., 2004).
2.4. Preparation, storage and transferring conditions
In airborne virus sampling with impinger, the samples are prepared for PCR analysis. In Faridi et al. study, the liquid impinger containing a mixture containing DMEM, streptomycin, penicillin, and anti-foam reagent isoamyl alcohol was ultra-centrifuged at 110,000 ×g for 1.5 h at 4 °C, followed by pellet suspending in the sampling medium and then, the collected samples were immediately transferred to a clinical virology laboratory (Faridi et al., 2020). In the use of MD-8 airscan sampling method, after sampling with PTFE filter, the filters were dissolved aseptically in 30 mL of viral transport medium. A mixture of sterile phosphate buffer with 10% fetal calf serum, 10,000 U/mL penicillin, 10 mg streptomycin, 25 μg amphotericin B has been introduced as a proper viral transport medium for trapped viruses. It should be noted that in air sampling with MD-8 airscan sampler, sampling conditions, such as relative humidity and temperature play key role on the collection performance of the used gelatin filter. In this regard, low relative humidity can cause desiccation of viruses, while high relative humidity can lead to dissolution of gelatin filters (Verreault et al., 2008). In Booth et al. study, on the side by side sampling with a wet air sampler (impinger like sampler) and PTEF filter cassette, the collected samples were shipped with ice packs and refrigerated upon receipt at the laboratory (Booth et al., 2005). In Chia et al. (2020) study, all samples were immediately stored at refrigerator (4 °C) and then kept at −80 °C per unless immediately analyzed. According to the previous studies, the air sample should be stored at 4 °C after sampling and during shipment, and then stored - 80 °C until viral or molecular biological analysis. Storage at −80 °C is not required when immediate PCR analysis is performed. In Kim et al. study, all air samples were collected with a MD-8 airscan sampler and then filtered through 0.1-μm pore syringe filter units to remove bacterial contamination prior to PCR analysis. Then, the filtered samples were incubated with Vero E6 in DMEM with 100 IU/mL penicillin and 100 μg/mL streptomycin at 37 °C in a CO2 incubator. After 14 days, the culture supernatants and lysates of Vero E6 cells were harvested and then used for RT-PCR analysis (Kim et al., 2016).
3. Identification methods
Present methods for detecting SARS-CoV-2 included RT-PCR and reverse transcription-loop-mediated isothermal amplification (RT-LAMP). Currently, RT-PCR is considered the gold standard for the detection of SARS-CoV-2 due to its many advantages such as high sensitivity and ease of use. However, it has been reported that the positive rate of RT-qPCR assay of pharyngeal swab samples is in the range 30–60%. Until now, the RT-PCR technique has been commonly used for the determination of SARS-CoV-2 in various matrices such as saliva, blood, urine, stool, surfaces, sputum and air (Peng et al., 2020; Wang et al., 2020). It has been reported that the RT-PCR is able to detect 4–8 copies of virus with a 95% confidence interval. In most of the reviewed studies, PCR analysis has been used for detecting corona viruses in air samples. Real-time PCR and related methods have revolutionized the laboratory diagnosis of viral respiratory infections because of their high detection sensitivity, rapidness and potential for simultaneous detection of 15 or more respiratory agents (Olofsson et al., 2011). In this regard, in Faridi et al. study, a Vazyme Viral RNA/DNA Mini Kit was used for extracting the viral RNA from sample matrices and then kept in an elution buffer. Finally, 5 mL of prepared RNA sample was used for RT-PCR analysis (Faridi et al., 2020). In Azhar et al. study on the detection of MERS-Cov virus in air, RT-PCR was applied for detection of the target virus. For this reason, RNA was extracted from the dissolved filter solution using a viral RNA minikit. Eluted RNA was tested by real-time RT-PCR analysis (Azhar et al., 2014). Most of the reviewed studies have used RT-PCR, which confirmed its enough sensitivity for detecting SARS-Cov 2 virus in air.
A recent study has used droplet digital PCR (ddPCR) with a high limit of detection (LOD) to detect SARS-CoV-2 in swab samples. The limit of blanks (LoBs) of these dPCR methods was reported to be 1.6, 1.6 and 0.8 copies/reaction for ORF 1ab, N and E gene, respectively. Moreover, its LOD was 2 copies/reaction, which considered as a high sensitivity. The overall accuracy of the ddPCR was 95.5% (Dong et al., 2020). However, this method has not yet been used for detection of corona viruses in air to be addressed and discussed in this review. Further studies are required to evaluate its performance in detecting such viruses in air. Moreover, Seo et al. introduced a sensor-based method for rapid detection of SARS-Cov-2 that may be useful for air sample as well. They used a field-effect transistor (FET)-based biosensing device for detecting these viral clinical samples, and in order to produce the sensor, graphene sheets of the FET was coated with a specific antibody against SARS-CoV-2 spike protein. The performance of the introduced method by Seo et al., was tested for nasopharyngeal swab samples of COVID-19 patients. It has been reported that the sensor-based method offered many advantages over the conventional detection methods such as high sensitivity and requiring no sample pretreatment (Seo et al., 2020). This detection method has the potential to detect low load of SARS-Cov-2 in air due to its high sensitivity. To the best of our knowledge, no study has used this method for determination of SARS-Cov-2 in air samples. Also, LAMP as a novel nucleic acid amplification method has been recently used for the rapid detection of SARS-CoV in clinical samples. This method is based on the amplification of nucleic acid, which offers many advantages such as high accuracy, high selectivity, and high speed detection performance under isothermal conditions. Another advantage of this technique is related to its ability to quantify the viral load with a simple method. The LAMP method provides an extremely high amplification efficiency because of its isothermal conditions, which there is no time loss for changing temperature. However, LAMP method could have higher rates of false positives. In this novel method, the nucleic acid amplification is associated with the production of magnesium pyrophosphate. The produced magnesium pyrophosphate causes turbidity that can be directly measured using an inexpensive turbidity meter (Hong et al., 2004). Until now, no study has evaluated the performance of the LAMP technique for detection SARS-Cov virus in air. However, Huang et al. used RT-LAMP for SARS-Cov-2 detection in clinical samples and introduced it as a sensitive, rapid and simple method that can be used even with naked eye. It has been reported that the RT-LAMP reaction can be conducted at 20 min and a constant temperature of 65 °C. The LOD of this method was reported to be 80 copies of viral RNA per mL sample (Huang et al., 2020), which is less than that of PCR method.
TEM is also a useful method for detection and characterization of virus in different matrices. The TEM technique provides an immediate overview of actual status, morphological characteristics viruses. In this method, as a first step in pathogen recognition, it is necessary to provide a sample containing a high viral load. For this reason, in most case, it is required to culture the virus in the sample matrix with a special culture medium. Since the TEM technique targets proteins, it is unbiased against RNA or DNA genomes. In this regard, observing the virus morphology allows the operator to immediate preliminary classify into family level based on its shape, size, structure and stability (King et al., 2011; Richert-Pöggeler et al., 2019). An accurate and detailed protocol for preparation and determination of a kind of corona virus (MERS-Cov) by RT-PCR and TEM has been described in Kim et al. study, which reported some PCR positive air samples for MERS-Cov (Kim et al., 2016). Briefly, in the mentioned study, the air collected samples with MD-8 airscan sampler were filtered through 0.1-μm pore syringe filter units for minimizing bacterial contamination, cultured Vero E6 cells in DMEM containing 100 IU/mL penicillin and 100 μg/mL streptomycin at 37 °C, and eventually the culture supernatants and lysates of Vero E6 cells was collected for RT-PCR analysis. After that, the harvested cells were centrifuged for 3 min at 1000 rpm to remove cellular debris. The pellets were re-suspended in washing buffer containing 0.1 M phosphate buffer and then centrifuged at 3000 rpm for 5 min. After thoroughly removing washing buffer, the cells were fixed with 2.5% glutaraldehyde at 4 °C overnight and characterized using TEM analysis (Kim et al., 2016). Moreover, in Ong et al. study, after sampling with MD-8 airscan sampler and 37-mm filter cassettes and 0.3-μm PTFE filter, specific real-time RT-PCR targeting RNA-dependent RNA polymerase and E genes was used for detection of SARS-CoV-2 in airborne infection isolation rooms (Ong et al., 2020). In Agranovski et al. study, TEM technique was used for the determination of SARS virus (SARS-Cov) in air samples. For this purpose, the trapped SARS virus in the proposed sampler method consisted of a HEPA filters installed in the pipeline connecting sampler and vacuum pump was inoculated onto confluent monolayers of Vero cells in flasks and then incubated at 37 °C for 60 min. Afterwards, the Inoculated cell cultures were stored with RPMI 1640 supplemented with 1% fetal bovine serum, and then the culture supernatant was used for investigation of the presence of the target virus by TEM technique. Formvar coated TEM grids were added into the cell culture supernatant for 7 min. Negative staining was carried out with 2% phosphotungstic acid (pH 7.4) for 7 min, PTA (pH 6.0) for 1–7 min, 2% ammonium molybdate (AMo, pH 6.5) for 1 min, 2% methylamine tungstate (pH 5.8) for 1 min and 1% aqueous uranyl acetate (UAc) for 5–45 s. The collected sampled were analyzed using a TEM analyzer at magnification range 10,000–60,000 (Agranovski et al., 2004). In Guo et al. study, RT-PCR was applied for detection of SARS-Cov-2 in hospital indoor air, which reported some positive air samples in near air outlets, patients' rooms, and in doctors' office area (Guo et al., 2020). It seems that the selection of sampler and sampling volume (by adjusting sampling flow rate and sampling time) play a key role in the detection of virus in air. By reviewing the published papers, it can be said that the corona viruses including, SARS-Cov, MERS-Cov and SARS-Cov-2 have been only detected in air samples that have been taken with a sampler at a high sampling volume. For example, in Guo et al. study, reported PCR positive for SARS-Cov-2 using a SASS 2300 Wetted Wall Cyclone Sampler with a sampling volume of 9000 L (Guo et al., 2020). In Kim et al., and Agranovski et al., studies, some air samples were positive for MERS-Cov and SARS-Cov viruses, respectively, which in both of them, the sampling volume was about 1000 L (Agranovski et al., 2004). In Liu et al. study sterilized gelatin filters with pore size of 3 μm placed in styrene filter cassette was utilized for indoor air sampling of SARS-Cov-2 with a flow rate of 5 L/min for 1 h, which reported a very low concentration of SARS-CoV-2 RNA in aerosols in isolation wards and ventilated patient rooms and a higher concentration of this virus in the toilet areas used by the patients (Liu et al., 2020).
4. Conclusion
This manuscript aimed at reviewing published papers for sampling and detection of SARS-Cov-2 as a global health concern. By reviewing the published papers, it was found that SARS-Cov 2 was present in some air samples that were collected from patient's rooms in hospitals. However, due to the fact that in the most reviewed studies, sampling was performed in the patient's room, it seems difficult to discriminate whether it is airborne or transmitted through respiratory droplets. About the sampling method, most of the used samplers showed suitable performance for trapping SARS-Co and MERS-Cov viruses followed by PCR analysis. Moreover, some other influencing factors such as patient distance from the sampler, having a protective or oxygen masks, patient activity, coughing and sneezing during sampling time, air movement, air conditioning, patient density in the sampling site, temperature and humidity, sampler type, sampling conditions, storage and transferring conditions, and detection method, can affect the results. Most studies with positive test results have used gelatin filters, PTFE filters and cyclones, and no positive test result was observed in the use of impinger like sampler for SARS like virus. It seems necessary to consider a high sampling volume for trapping SARS-Cov-viruses in contaminated air. RT-PCR and TEM were commonly used for the detection of SARS-like viruses. DdPCR, FET-based biosensing and LAMP showed the potential to be used for sensitive detection of corona virus in clinical samples, and hence further studies are required to investigate their performance for sensitive and accurate detection of SARS-Cov-2 viruses in air.
CRediT authorship contribution statement
Ali Reza Rahmani: Conceptualization, Supervision. Mostafa Leili: Writing - review & editing. Ghasem Azarian: Conceptualization, Investigation. Ali Poormohammadi: Conceptualization, Writing - original draft.
Declaration of competing interest
The authors declared no conflict of interest regarding this paper.
References
- Agranovski I.E., Safatov A.S., Pyankov O.V., Sergeev A.N., Agafonov A.P., Ignatiev G.M., Ryabchikova E.I., Borodulin A.I., Sergeev A.A., Doerr H.W., Rabenau H.F., Agranovski V. Monitoring of viable airborne SARS virus in ambient air. Atmos. Environ. 2004;38:3879–3884. doi: 10.1016/j.atmosenv.2004.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., Hashem A.M., El-Kafrawy S.A., Sohrab S.S., Aburizaiza A.S., Farraj S.A., Hassan A.M., Al-Saeed M.S., Jamjoom G.A., Madani T.A. Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. MBio. 2014;5(4) doi: 10.1128/mBio.01450-14. (e01450-01414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs C.B. The airborne transmission of infection in hospital buildings: fact or fiction? Indoor Built Environ. 2003;12:9–18. doi: 10.1177/1420326X03012001002. [DOI] [Google Scholar]
- Blachere F.M., Cao G., Lindsley W.G., Noti J.D., Beezhold D.H. Enhanced detection of infectious airborne influenza virus. J. Virol. Methods. 2011;176:120–124. doi: 10.1016/j.jviromet.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Booth T.F., Kournikakis B., Bastien N., Ho J., Kobasa D., Stadnyk L., Li Y., Spence M., Paton S., Henry B. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 2005;191(9):1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton N.C., Grinshpun S.A., Reponen T. Physical collection efficiency of filter materials for bacteria and viruses. Ann. Occup. Hyg. 2007;51:143–151. doi: 10.1093/annhyg/mel073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V., Egloff M.-P., Longhi S., Ferron F., Rancurel C., Salomoni A., Durousseau C., Tocque F., Brémond N., Dobbe J.C. Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr. D Biol. Crystallogr. 2003;59(9):1628–1631. doi: 10.1107/S0907444903016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Sutjipto S., Lee P.H., Young B.E., Milton D.K. 2020. Detection of Air and Surface Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Hospital Rooms of Infected Patients. medRxiv. [DOI] [Google Scholar]
- Chinn R.Y., Sehulster L. 2003. Guidelines for Environmental Infection Control in Health-Care Facilities; Recommendations of CDC and Healthcare Infection Control Practices Advisory Committee (HICPAC) [PubMed] [Google Scholar]
- Chow E.J., Mermel L.A. Hospital-acquired respiratory viral infections: incidence, morbidity, and mortality in pediatric and adult patients. Open Forum Infect. Dis. 2017;4(1) doi: 10.1093/ofid/ofx006. (ofx006-ofx006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mbareche H., Lindsley W.G., Duchaine C. Field sampling of indoor bioaerosols. Aerosol Sci. Tech. 2020;54:572–584. doi: 10.1080/02786826.2019.1688759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon H.K., Heinsohn P.A., Miller J.D. AIHA; 2005. Field Guide for the Determination of Biological Contaminants in Environmental Samples. [Google Scholar]
- Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M. 2020. Highly Accurate and Sensitive Diagnostic Detection of SARS-CoV-2 by Digital PCR. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fabian P., McDevitt J.J., Houseman E.A., Milton D.K. Airborne influenza virus detection with four aerosol samplers using molecular and infectivity assays: considerations for a new infectious virus aerosol sampler. Indoor Air. 2009;19(5):433–441. doi: 10.1111/j.1600-0668.2009.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi S., Niazi S., Sadeghi K., Naddafi K., Yavarian J., Shamsipour M., Jandaghi N.Z.S., Sadeghniiat K., Nabizadeh R., Yunesian M. A field indoor air measurement of SARS-CoV-2 in the patient rooms of the largest hospital in Iran. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.-D., Wang Z.-Y., Zhang S.-F., Li X., Li L., Li C., Cui Y., Fu R.-B., Dong Y.-Z., Chi X.-Y. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T.C.T., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020 doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F.B. Transport of viral specimens. Clin. Microbiol. Rev. 1990;3:120–131. doi: 10.1128/cmr.3.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.W., Ramakrishnan M., Raynor P.C., Goyal S.M. Effects of humidity and other factors on the generation and sampling of a coronavirus aerosol. Aerobiologia. 2007;23:239–248. doi: 10.1007/s10453-007-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Chang S.Y., Sung M., Park J.H., Bin Kim H., Lee H., Choi J.-P., Choi W.S., Min J.-Y. Extensive viable Middle East respiratory syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Rev. Infect. Dis. 2016;63(3):363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M., Lefkowitz E., Adams M.J., Carstens E.B. Elsevier; 2011. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- Kouni S., Karakitsos P., Chranioti A., Theodoridou M., Chrousos G., Michos A. Evaluation of viral co-infections in hospitalized and non-hospitalized children with respiratory infections using microarrays. Clin. Microbiol. Infect. 2013;19:772–777. doi: 10.1111/1469-0691.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung M., Chan A.H. Control and management of hospital indoor air quality. Med. Sci. Monit. 2006;12(3):SR17–SR23. [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lindsley W.G., Green B.J., Blachere F.M., Martin S.B., Law B., Jensen P., Schafer M. 5th ed. National Institute for Occupational Safety and Health; Cincinnati (OH): 2017. Sampling and Characterization of Bioaerosols. NIOSH Manual of Analytical Methods. [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020:1–6. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Meiklejohn G., Kempe C., Downie A., Berge T., Vincent L.S., Rao A. Air sampling to recover variola virus in the environment of a smallpox hospital. Bull. World Health Organ. 1961;25(1):63. [PMC free article] [PubMed] [Google Scholar]
- NIOSH . NIOSH Manual of Analytical Methods (NMAM) fourth edition. 1998. BIOAEROSOL SAMPLING (indoor air) 0800, culturable organisms: bacteria, fungi, thermophilic actinomycetes; p. 1. [Google Scholar]
- Olofsson S., Brittain-Long R., Andersson L.M., Westin J., Lindh M. PCR for detection of respiratory viruses: seasonal variations of virus infections. Expert Rev. Anti Infect. 2011;9(8):615–626. doi: 10.1586/eri.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. Jama. 2020 doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Lednicky J.A., Wu C.Y. Collection, particle sizing and detection of airborne viruses. J. Appl. Microbiol. 2019;127(6):1596–1611. doi: 10.1111/jam.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Deng K., Lin B., Gao Z. 2020. 2019 Novel Coronavirus Can Be Detected in Urine, Blood, Anal Swabs and Oropharyngeal Swabs Samples, medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert-Pöggeler K.R., Franzke K., Hipp K., Kleespies R.G. Electron microscopy methods for virus diagnosis and high resolution analysis of viruses. Front. Microbiol. 2019;9:3255. doi: 10.3389/fmicb.2018.03255.eCollection. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandle T. In: Pharmaceutical Microbiology. Sandle T., editor. Woodhead Publishing; Oxford: 2016. 16 - Cleanrooms and environmental monitoring; pp. 199–217. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shi T., McLean K., Campbell H., Nair H. Aetiological role of common respiratory viruses in acute lower respiratory infections in children under five years: a systematic review and meta–analysis. J. Glob. Health. 2015;5(1) doi: 10.7189/jogh.05.010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeder M.C., Fackler J.C. Hospital-acquired viral infection increases mortality in children with severe viral respiratory infection. Pediatr. Crit. Care Med. 2011;12(6):e317–e321. doi: 10.1097/PCC.0b013e3182230f6e. [DOI] [PubMed] [Google Scholar]
- Verreault D., Moineau S., Duchaine C. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 2008;72(3):413–444. doi: 10.1128/MMBR.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Aarmink A., Wang W., Fabri T., PW G.K., de Jong M.C. Airborne virus sampling-efficiencies of samplers and their detection limits for infectious bursal disease virus (IBDV) Ann. Agric. Environ. Med. 2014;21(3) doi: 10.5604/12321966.1120585. [DOI] [PubMed] [Google Scholar]