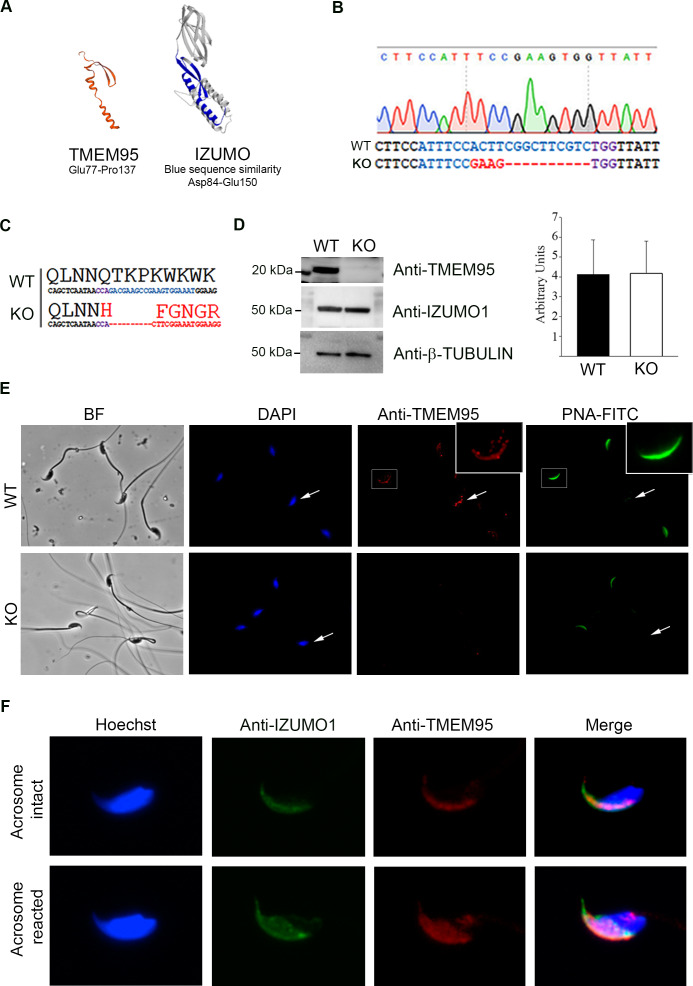
Figure 1. Generation of TMEM95 deficient mice.
(A) Structure prediction of TMEM95 protein (left) using IZUMO1 (right) as template, created by SWISS-MODEL software. (B) Tmem95 KO allele generated following CRISPR-mediated edition. CRISPR target sequence and PAM are depicted in blue and purple letters, respectively. (C) The deletion of 10 bp altered Tmem95 ORF. Large letters indicate the aminoacid sequence corresponding to the codons (DNA sequence) shown in smaller letters below. (D) Western Blot images for TMEM95, IZUMO1 and β-tubulin proteins from protein extracts from WT or KO sperm. Graph on right indicates the abundance of IZUMO1 in WT and KO extracts. (E) Immunocytochemistry images of KO and WT sperm stained with an antibody against TMEM95 and the acrosomal stain PNA. TMEM95 localized to the acrosomal cap in acrosome intact sperm and in the equatorial segment after acrosome reaction. (F) Immunocytochemistry images of acrosome intact (upper images) or reacted (lower images) WT sperm stained against IZUMO1 and TMEM95. Both proteins relocalize to the equatorial segment following acrosome reaction.
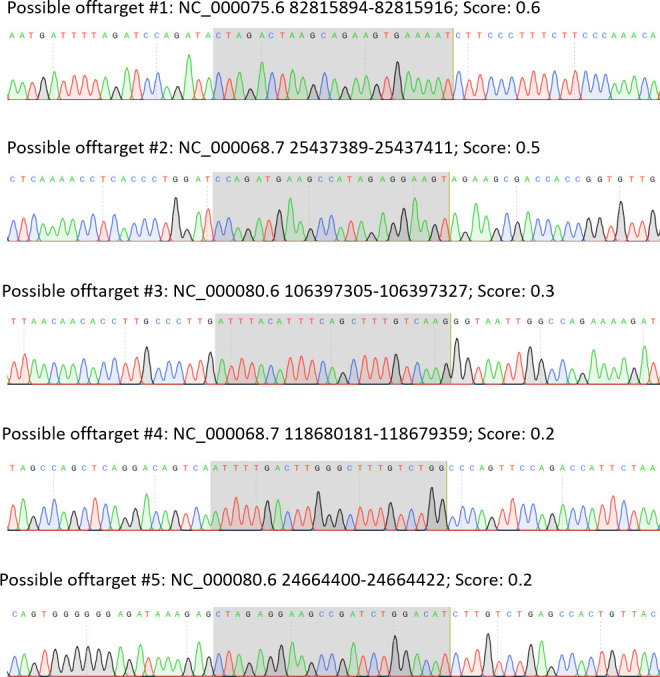
Figure 1—figure supplement 1. Offtarget analysis.