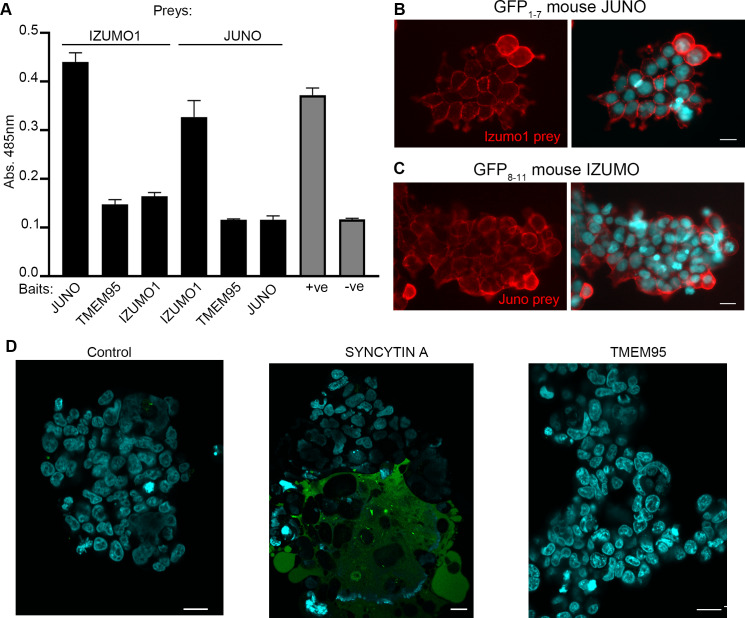
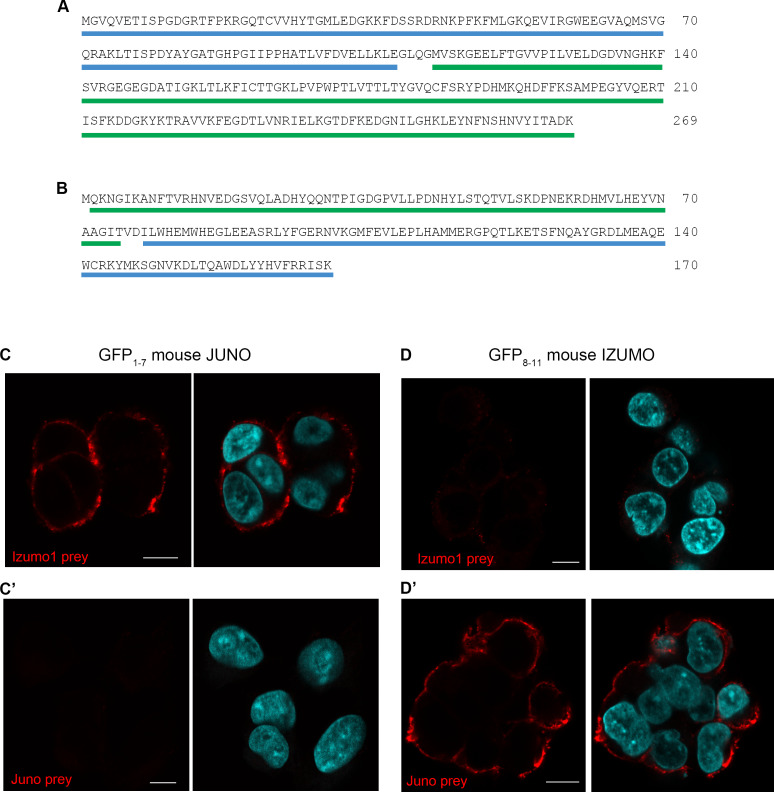
Figure 3. TMEM95 does not interact with JUNO nor IZUMO1.
(A) Binding analysis using the AVEXIS assays shows that the soluble recombinant TMEM95 ectodomain does not interact with JUNO nor with IZUMO1. The entire ectodomains of the named proteins were expressed in HEK293-6E cells either as biotinylated baits or as pentameric beta-lactamase-tagged preys. Bait proteins were immobilised on streptavidin-coated plates and captured prey proteins quantified by measuring the absorbance of a colorimetric reaction product of the beta-lactamase substrate, nitrocefin. The CD200R (bait)-CD200 (prey) binding pair was used as positive control. The same prey, CD200R, was tested against TMEM95 and is shown as negative control. Bars represent means + s.d.; n = 3. (B) HEK293 cells stably expressing the N-terminal half of GFP (GFP1-7) and mouse JUNO stained with a highly avid IZUMO1 probe. (C) HEK293 cells stably expressing the C-terminal half of GFP (GFP8-11) and mouse IZUMO1 stained with a highly avid JUNO. (D) TMEM95 does not induce fusion when expressed in HEK293T cells in the presence of JUNO and IZUMO1 using a GFP-complementation cell fusion assay. HEK293T cells expressing either half of GFP and either JUNO or IZUMO1 were mixed and their fusogenic ability visualized by GFP fluorescence. The IZUMOI-expressing cells were either mock transfected prior to mixing (Control), transfected with Syncitin a, as a positive fusion control, or Tmem95. By contrast to the cells transfected with Syncytin a, Tmem95 did not induce cell fusion. Cell nuclei are stained with DAPI and scale bar represents 20 µm.