FIGURE 1.
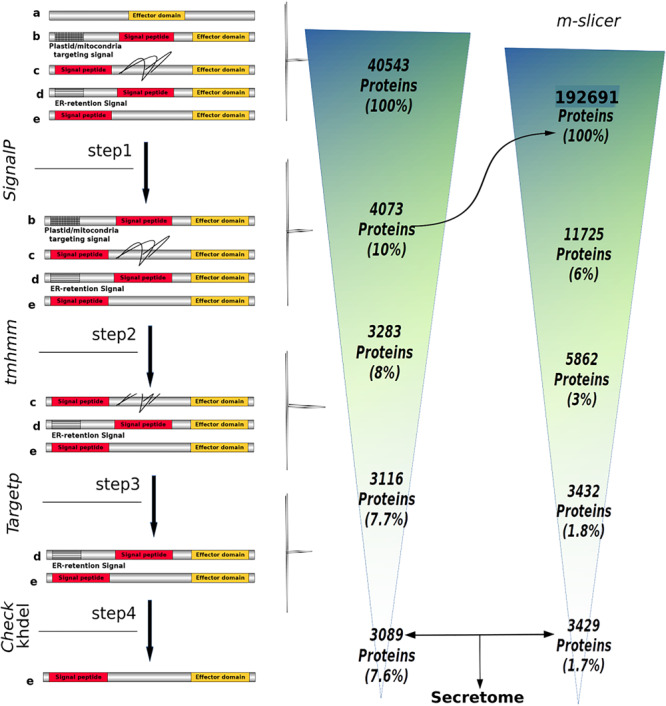
Number of proteins obtained throughout the SecretSanta pipeline used for the prediction of the Phytophthora betacei secretome. The secretome prediction pipeline is depicted on the left. The proteins include: (a) proteins without signal peptide, (b) proteins with mitochondria/plastid targeting signals, (c) proteins with transmembrane domains (black lines) located after signal peptide, (d) proteins with short ER lumen retention signals, and (e) proteins with a signal peptide and an effector domain; ensuring the absence of motifs and domains preventing the protein from being secreted or those targeting it to specific organelles. Step 1: identification of short signal peptides at the N-terminal end of a protein using different version of SignalP. In steps 2, 3 and 4, proteins with motifs and domains preventing the protein from being secreted or those that target it to specific organelles are filtered, using TMHMM, TargetP and KHDEL. The number of proteins retained in each step is shown on the right. The m_slicer function that generates sequences with alternative translation start sites, had as input the first resulting set of predicted proteins having a signal peptide (Rightward arrow).
