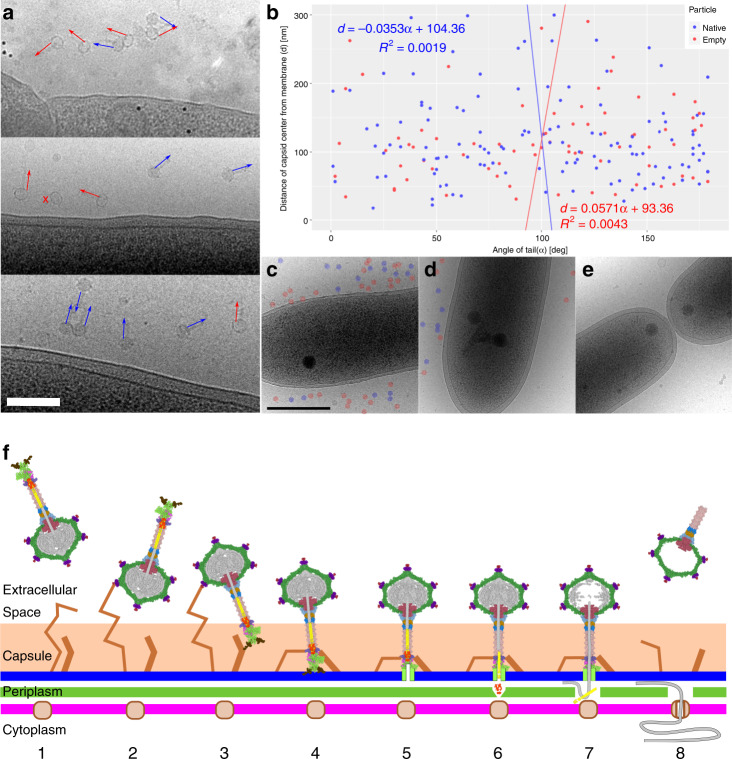
Fig. 6. Mechanism of DNA delivery by RcGTA.
a, b RcGTA particles attach to cells in random orientations. a Cryo-electron micrographs of particles of RcGTA attached to cells of R. capsulatus. The panel includes images from three biological replicates. Orientations of RcGTA tails are highlighted with arrows. Blue indicates genome-containing particles and red empty ones. A cross next to a particle indicates that its tail is not visible in the projection image. Black dots are fiducial markers. Scale bar represents 200 nm. b Distribution of orientations of tails of RcGTA particles (x-axis) versus distance of capsid center from outer cell membrane (y-axis). The tail orientation has values from 0°, when the tail points at the membrane, to 180°, when the tail points away from membrane. The particles are oriented randomly. c–e Cells of R. capsulatus are heterogeneous in their capacity to bind RcGTA. Electron micrographs of three R. capsulatus cells from the same experiment using multiplicity of Rif-transferring RcGTA of 0.0002. Some cells were covered with numerous RcGTA particles (c), some had tens of them attached (d), whereas the remaining ones only attracted a few (e). Native and empty RcGTA particles are highlighted with blue and red circles, respectively. f Model of RcGTA-mediated DNA delivery. (1) Free particle. (2) RcGTA attaches to the cell capsule by the head fibers. (3) Particle reorients by the binding of tail fibers to outer membrane receptors. (4) Particle attaches to the membrane by putative receptor-binding domains of the baseplate. (5) Penetration of the outer membrane by iris/penetration domain of megatron protein. (6) Ejection of cell-wall peptidase into periplasm enables degradation of cell wall. (7) Ejection of tape measure protein with DNA to periplasmic space. (8) Uptake of DNA by cell competence system.