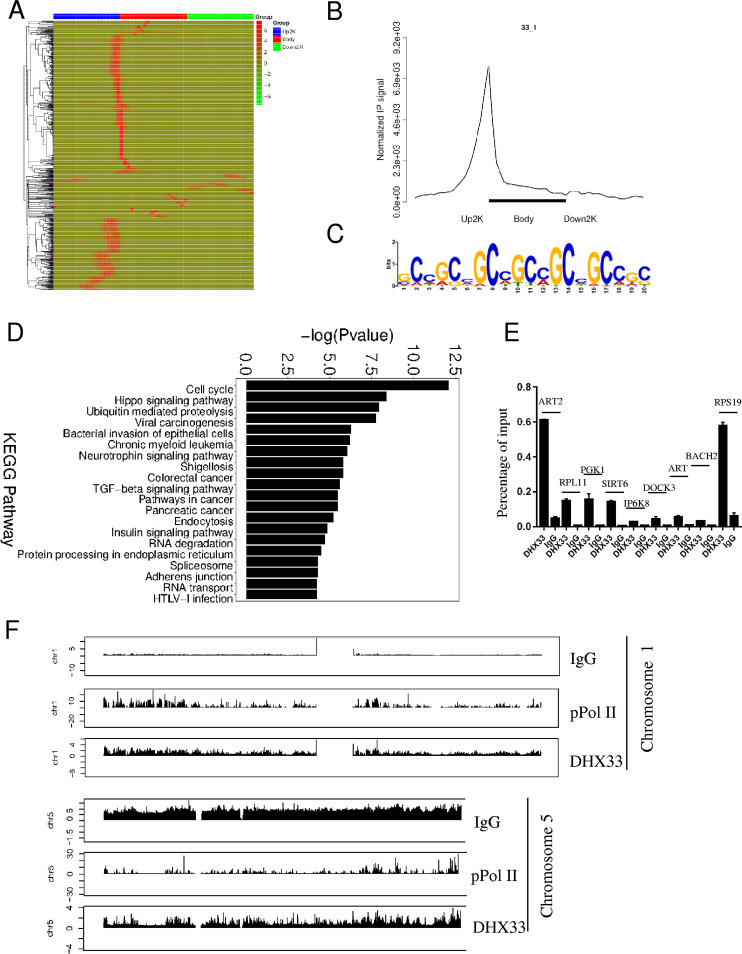
FIG 2.
DHX33 protein bound to the promoters of a subset of genes. (A) H1299 cells were infected by lentivirus encoding wild-type DHX33. Five days postinfection, cells were subjected to ChIP-seq analysis with anti-DHX33 antibody; IgG served as a negative control, and anti-pPol II served as a positive control. The heat map shows the binding region (red) of DHX33 on different gene domains. The scale bar is shown on the right. (B) Normalized IP signal analysis for DHX33 on gene regions. (C) Motif analysis for DHX33 binding sites on genes. The most preferential motif is shown to be a CG-rich region. (D) KEGG pathway analysis of genes associated with DHX33 protein. Most fall into the categories of cell cycle, ubiquitin-mediated proteolysis, and cancer-related pathways. TGF-beta, transforming growth factor beta; HTLV-1, human T-cell leukemia virus type 1. (E) To validate the results for ChIP sequencing, quantitative PCR was conducted for pooled DNA fragments from the ChIP analysis performed as described for panel A. Nine representative genes were selected from the gene list. Enrichment signals are plotted as the input percentage. All demonstrated significant difference compared to the IgG control. (F) Peak distribution analysis for ChIP signals from IgG, anti-pPol II, and anti-DHX33 antibody. Results from two representative chromosomes are shown. The data for the whole genome can be found in the supplemental material. y-axis data represent the fold enrichment values.