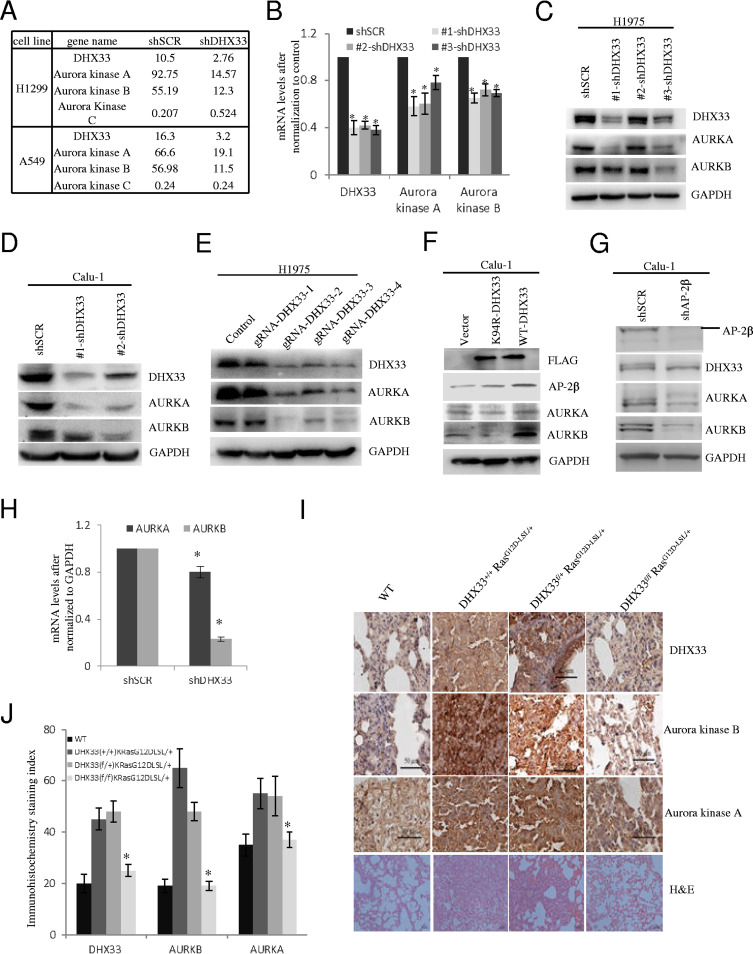
FIG 4.
DHX33 promoted gene transcription of Aurora kinase A/B. (A) RNA-seq was performed on H1299 or A549 cells after DHX33 was knocked down. The expression levels of Aurora kinase are shown. Both Aurora kinase A and Aurora kinase B were downregulated, while Aurora kinase C was either not affected or slightly upregulated. (B) The mRNA levels of Aurora kinase A and B were analyzed by QPCR after DHX33 knockdown in H1975 cells. GAPDH was used for normalization. *, P < 0.05; n = 3. (C and D) H1975 (C) and Calu-1 (D) cells were infected by lentivirus encoding either shSCR or multiple shDHX33s. Four days postinfection, whole-cell extracts were prepared and subjected to Western blotting with the indicated antibodies. GAPDH was used as an internal control. (E) H1975 cells were transfected by plasmids encoding Cas9 and guide RNAs targeting different exons of DHX33 gene, with empty vector as a control. Two days posttransfection, whole-cell extracts were subjected to Western blotting with the indicated antibodies. GAPDH served as an internal control. (F) Calu-1 cells were infected with lentivirus encoding empty vector, the K94R helicase-defective DHX33 mutant, or wild-type DHX33. Four days postinfection, whole-cell extracts were subjected to Western blotting with the designated antibodies. (G) Calu-1 cells were infected with lentivirus encoding either shSCR or shAP-2β. Four days postinfection, whole-cell extracts were subjected to Western blotting with the designated antibodies. (H) Total RNA was extracted from Calu-1 cells in 4G and then subjected to reverse transcription-PCR (RT-PCR) analysis for the expression of Aurora kinase A and B. *, P < 0.05; n = 3. (I) Mice with the four different genotypes were treated with adenovirus encoding Cre recombinase according to a standard protocol (28). Lung tissues from the four designated groups were analyzed 2 months later; immunohistochemistry staining was performed with the indicated antibody for gene expression analysis. Tissues were then counterstained with hematoxylin (blue). Brownish coloring indicates positive staining. Hematoxylin and eosin (H&E) staining results are also shown at the bottom. (J) Quantitation of the immunohistochemistry (IHC) signals. Five representative areas were selected for evaluation of IHC signals. *, P < 0.05; n = 5.